Sorting data set 20010214 tetrode B (channels 2, 3, 5, 7)
Table of Contents
- 1. Introduction
- 2. Tetrode B (channels 2, 3, 5, 7) analysis
- 2.1. Loading the data
- 2.2. Five number summary
- 2.3. Plot the data
- 2.4. Data normalization
- 2.5. Spike detection
- 2.6. Cuts
- 2.7. Events
- 2.8. Removing obvious superposition
- 2.9. Dimension reduction
- 2.10. Exporting for
GGobi - 2.11. kmeans clustering with 10
- 2.12. Long cuts creation
- 2.13. Peeling
- 2.14. Getting the spike trains
- 2.15. Getting the inter spike intervals and the forward and backward recurrence times
- 2.16. Making "all at once"
- 3. Analyzing a sequence of trials
- 4. Systematic analysis of the 30 trials from
Spontaneous_2backwards - 5. 30 trials of
Spontaenous_1backwards - 6. 25 trials with
Cis-3-Hexen-1-ol(C3H_1) - 7. 25 trials with
Citral - 8. 25 trials with
Vanilla - 9. 25 trials with
Octanol - 10. 25 trials with
Mint - 11. 25 trials with
C3H_2 - 12. 30 trials of
Spontaneous_3 - 13. 30 trials of
Spontaneous_4 - 14. 30 trials of
C3H_3 - 15. 10 trials of
C3H_4 - 16. 30 trials of
C3H_5 - 17. 30 trials of
C3H_6
1 Introduction
This is the description of how to do the (spike) sorting of tetrode B (channels 2, 3, 5, 7) from data set locust20010214.
1.1 Getting the data
The data are in file locust20010214.hdf5 located on zenodo and can be downloaded interactivelly with a web browser or by typing at the command line:
wget https://zenodo.org/record/21589/files/locust20010214_part1.hdf5 wget https://zenodo.org/record/21589/files/locust20010214_part2.hdf5
In the sequel I will assume that R has been started in the directory where the data were downloaded (in other words, the working direcory should be the one containing the data.
The data are in HDF5 format and the easiest way to get them into R is to install the rhdf5 package from Bioconductor. Once the installation is done, the library is loaded into R with:
library(rhdf5)
We can then get a (long and detailed) listing of our data file content with (result not shown):
h5ls("locust20010214_part1.hdf5")
We can get the content of LabBook metadata from the shell with:
h5dump -a "LabBook" locust20010214_part1.hdf5
1.2 Getting the code
The code can be sourced as follows:
source("https://raw.githubusercontent.com/christophe-pouzat/zenodo-locust-datasets-analysis/master/R_Sorting_Code/sorting_with_r.R")
A description of the functions contained in this file can be found at the following address: http://xtof.perso.math.cnrs.fr/locust.html.
2 Tetrode B (channels 2, 3, 5, 7) analysis
We now want to get our "model", that is a dictionnary of waveforms (one waveform per neuron and per recording site). To that end we are going to use the last 60 s of data contained in the Spontaneous_2 Group (in HDF5 jargon).
2.1 Loading the data
So we start by loading the data from channels 2, 3, 5, 7 into R:
lD = rbind(cbind(h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch02"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch03"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch05"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch07")))
dim(lD)
| 431548 |
| 4 |
2.2 Five number summary
We get the Five number summary with:
summary(lD,digits=2)
| Min. : 98 | Min. : 610 | Min. : 762 | Min. : 475 |
| 1st Qu.:2011 | 1st Qu.:2015 | 1st Qu.:2017 | 1st Qu.:2014 |
| Median :2058 | Median :2057 | Median :2057 | Median :2059 |
| Mean :2055 | Mean :2055 | Mean :2054 | Mean :2056 |
| 3rd Qu.:2105 | 3rd Qu.:2099 | 3rd Qu.:2095 | 3rd Qu.:2102 |
| Max. :3634 | Max. :3076 | Max. :2887 | Max. :3143 |
It shows that the channels have very similar properties as far as the median and the inter-quartile range (IQR) are concerned. The minimum is much smaller on the first channel. This suggests that the largest spikes are going to be found here (remember that spikes are going mainly downwards).
2.3 Plot the data
We "convert" the data matrix lD into a time series object with:
lD = ts(lD,start=0,freq=15e3)
We can then plot the whole data with (not shown since it makes a very figure):
plot(lD)
2.4 Data normalization
As always we normalize such that the median absolute deviation (MAD) becomes 1:
lD.mad = apply(lD,2,mad) lD = t((t(lD)-apply(lD,2,median))/lD.mad) lD = ts(lD,start=0,freq=15e3)
Once this is done we explore interactively the data with:
explore(lD,col=c("black","grey70"))
Most spikes can be seen on the 4 recording sites, there are many different spike waveform and the signal to noise ratio is really good!
2.5 Spike detection
Since the spikes are mainly going downwards, we will detect valleys instead of peaks:
lDf = -lD filter_length = 5 threshold_factor = 4 lDf = filter(lDf,rep(1,filter_length)/filter_length) lDf[is.na(lDf)] = 0 lDf.mad = apply(lDf,2,mad) lDf_mad_original = lDf.mad lDf = t(t(lDf)/lDf_mad_original) thrs = threshold_factor*c(1,1,1,1) bellow.thrs = t(t(lDf) < thrs) lDfr = lDf lDfr[bellow.thrs] = 0 remove(lDf) sp0 = peaks(apply(lDfr,1,sum),15) remove(lDfr) sp0
eventsPos object with indexes of 1950 events. Mean inter event interval: 221.2 sampling points, corresponding SD: 204.2 sampling points Smallest and largest inter event intervals: 16 and 2904 sampling points.
Every time a filter length / threshold combination is tried, the detection is checked interactively with:
explore(sp0,lD,col=c("black","grey50"))
2.6 Cuts
We proceed as usual to get the cut length right:
evts = mkEvents(sp0,lD,49,50)
evts.med = median(evts)
evts.mad = apply(evts,1,mad)
plot_range = range(c(evts.med,evts.mad))
plot(evts.med,type="n",ylab="Amplitude",
ylim=plot_range)
abline(v=seq(0,400,10),col="grey")
abline(h=c(0,1),col="grey")
lines(evts.med,lwd=2)
lines(evts.mad,col=2,lwd=2)
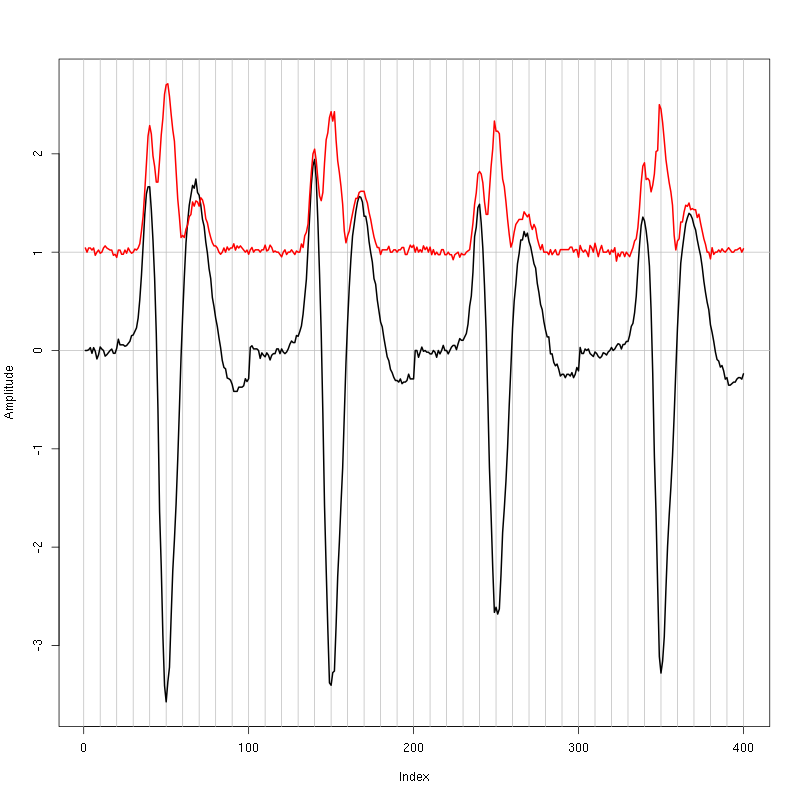
Figure 1: Setting the cut length for the data from tetrode B (channels 2, 3, 5, 7). We see that we need 20 points before the peak and 30 after.
We see that we need roughly 20 points before the peak and 30 after.
2.7 Events
We now cut our events:
evts = mkEvents(sp0,lD,19,30) summary(evts)
events object deriving from data set: lD. Events defined as cuts of 50 sampling points on each of the 4 recording sites. The 'reference' time of each event is located at point 20 of the cut. There are 1950 events in the object.
We can as usual visualize the first 200 events with:
evts[,1:200]
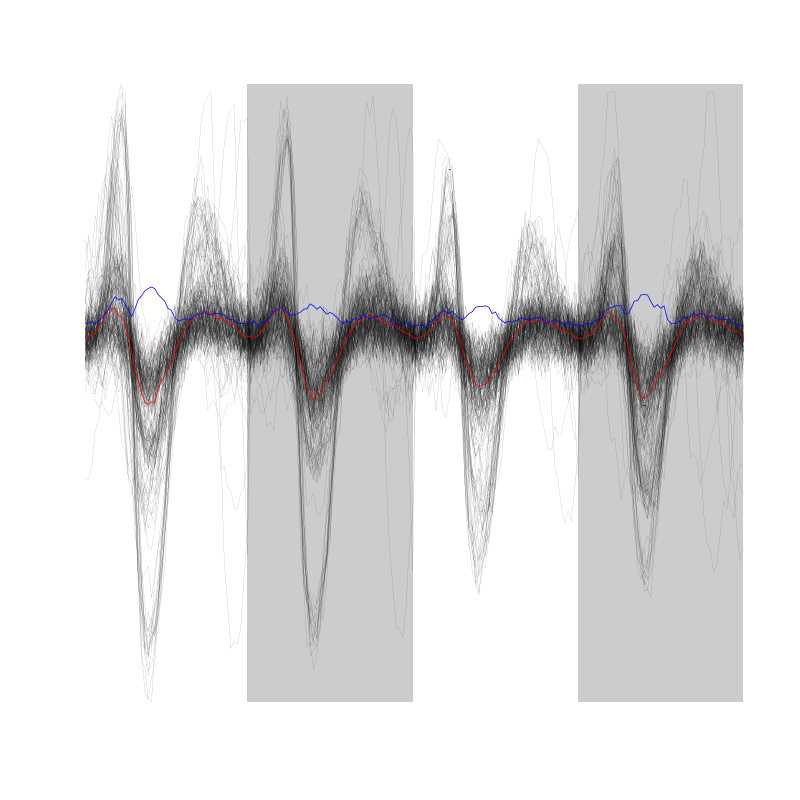
Figure 2: First 200 events for the data from tetrode B (channels 1, 3, 5, 7).
There are few superpositions so we try to remove the most obvious ones before doing the dimension reduction.
2.8 Removing obvious superposition
We define function goodEvtsFct with:
goodEvtsFct = function(samp,thr=3) {
samp.med = apply(samp,1,median)
samp.mad = apply(samp,1,mad)
below = samp.med < 0
samp.r = apply(samp,2,function(x) {x[below] = 0;x})
apply(samp.r,2,function(x) all(abs(x-samp.med) < thr*samp.mad))
}
We apply it with a threshold of 8 times the MAD:
goodEvts = goodEvtsFct(evts,8)
2.9 Dimension reduction
We do a PCA on our good events set:
evts.pc = prcomp(t(evts[,goodEvts]))
We look at the projections on the first 4 principle components:
panel.dens = function(x,...) {
usr = par("usr")
on.exit(par(usr))
par(usr = c(usr[1:2], 0, 1.5) )
d = density(x, adjust=0.5)
x = d$x
y = d$y
y = y/max(y)
lines(x, y, col="grey50", ...)
}
pairs(evts.pc$x[,1:4],pch=".",gap=0,diag.panel=panel.dens)
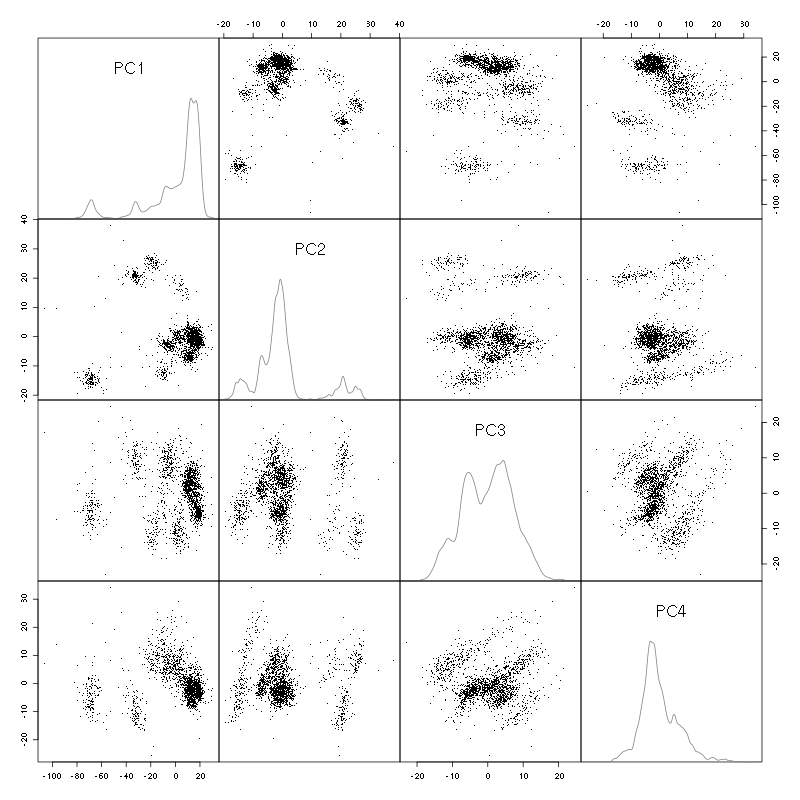
Figure 3: Events from tetrode B (channels 2, 3, 5, 7) projected onto the first 4 PCs.
I see at least 10 clusters. We can also look at the projections on the PC pairs defined by the next 4 PCs:
pairs(evts.pc$x[,5:8],pch=".",gap=0,diag.panel=panel.dens)
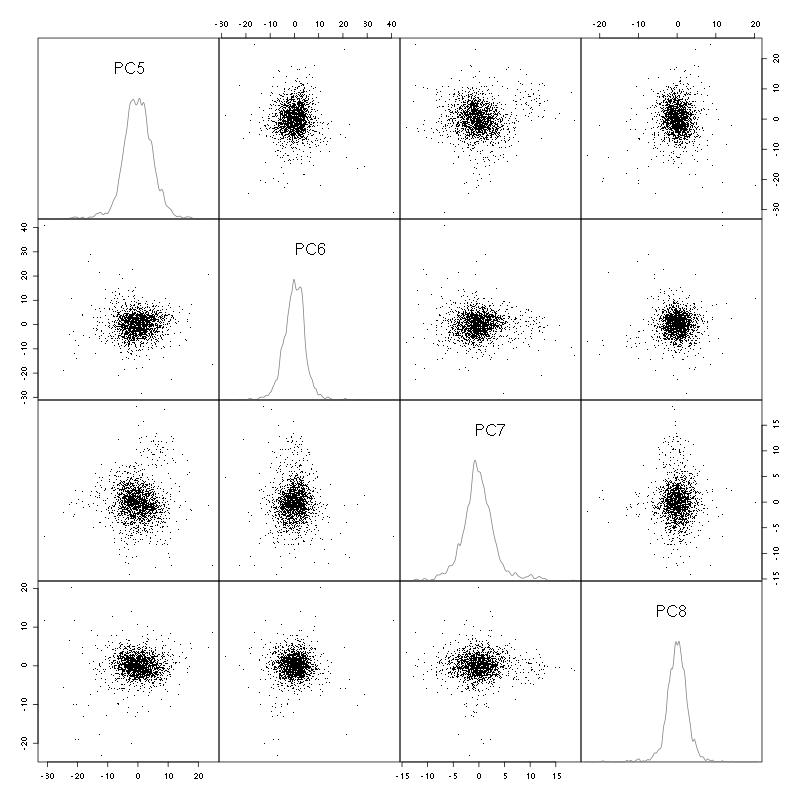
Figure 4: Events from tetrode B (channels 2, 3, 5, 7) projected onto PC 5 to 8.
There is not much structure left beyond the 4th PC.
2.10 Exporting for GGobi
We export the events projected onto the first 8 principle components in csv format:
write.csv(evts.pc$x[,1:8],file="tetB_evts.csv")
Using the rotation display of GGobi with the first 3 principle components and the 2D tour with the first 4 components I see at least 10. So we will start with a kmeans with 10 centers.
2.11 kmeans clustering with 10
nbc=10
set.seed(20110928,kind="Mersenne-Twister")
km = kmeans(evts.pc$x[,1:4],centers=nbc,iter.max=100,nstart=100)
label = km$cluster
cluster.med = sapply(1:nbc, function(cIdx) median(evts[,goodEvts][,label==cIdx]))
sizeC = sapply(1:nbc,function(cIdx) sum(abs(cluster.med[,cIdx])))
newOrder = sort.int(sizeC,decreasing=TRUE,index.return=TRUE)$ix
cluster.mad = sapply(1:nbc, function(cIdx) {ce = t(evts[,goodEvts]);ce = ce[label==cIdx,];apply(ce,2,mad)})
cluster.med = cluster.med[,newOrder]
cluster.mad = cluster.mad[,newOrder]
labelb = sapply(1:nbc, function(idx) (1:nbc)[newOrder==idx])[label]
We write a new csv file with the data and the labels:
write.csv(cbind(evts.pc$x[,1:4],labelb),file="tetB_sorted.csv")
It gives what was expected.
We get a plot showing the events attributed to each of the first 5 units with:
layout(matrix(1:5,nr=5)) par(mar=c(1,1,1,1)) for (i in (1:5)) plot(evts[,goodEvts][,labelb==i],y.bar=5)
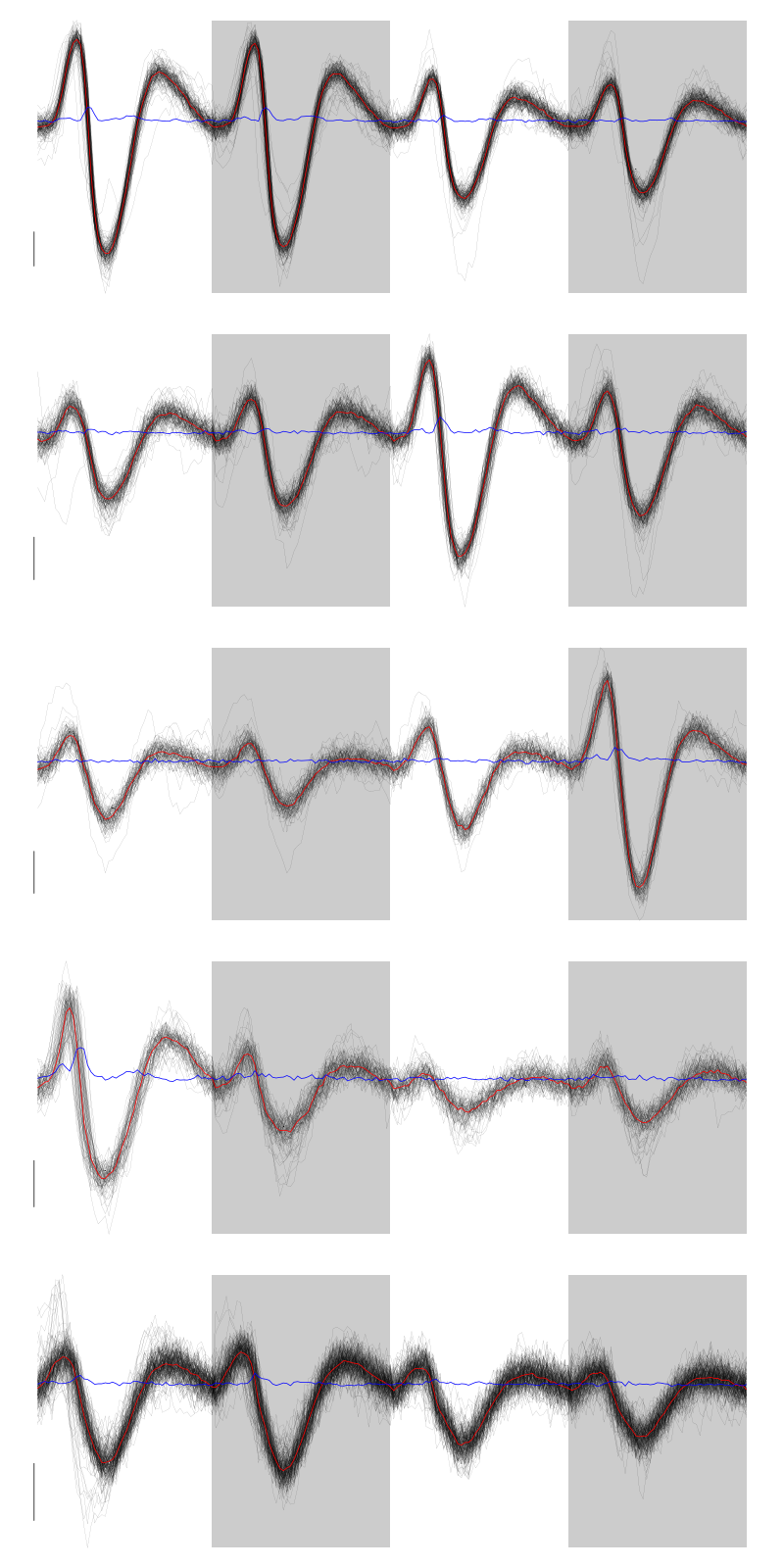
Figure 5: The events of the first five clusters of tetrode B
We get a plot showing the events attributed to each of the last 5 units with:
layout(matrix(1:5,nr=5)) par(mar=c(1,1,1,1)) for (i in (6:10)) plot(evts[,goodEvts][,labelb==i],y.bar=5)
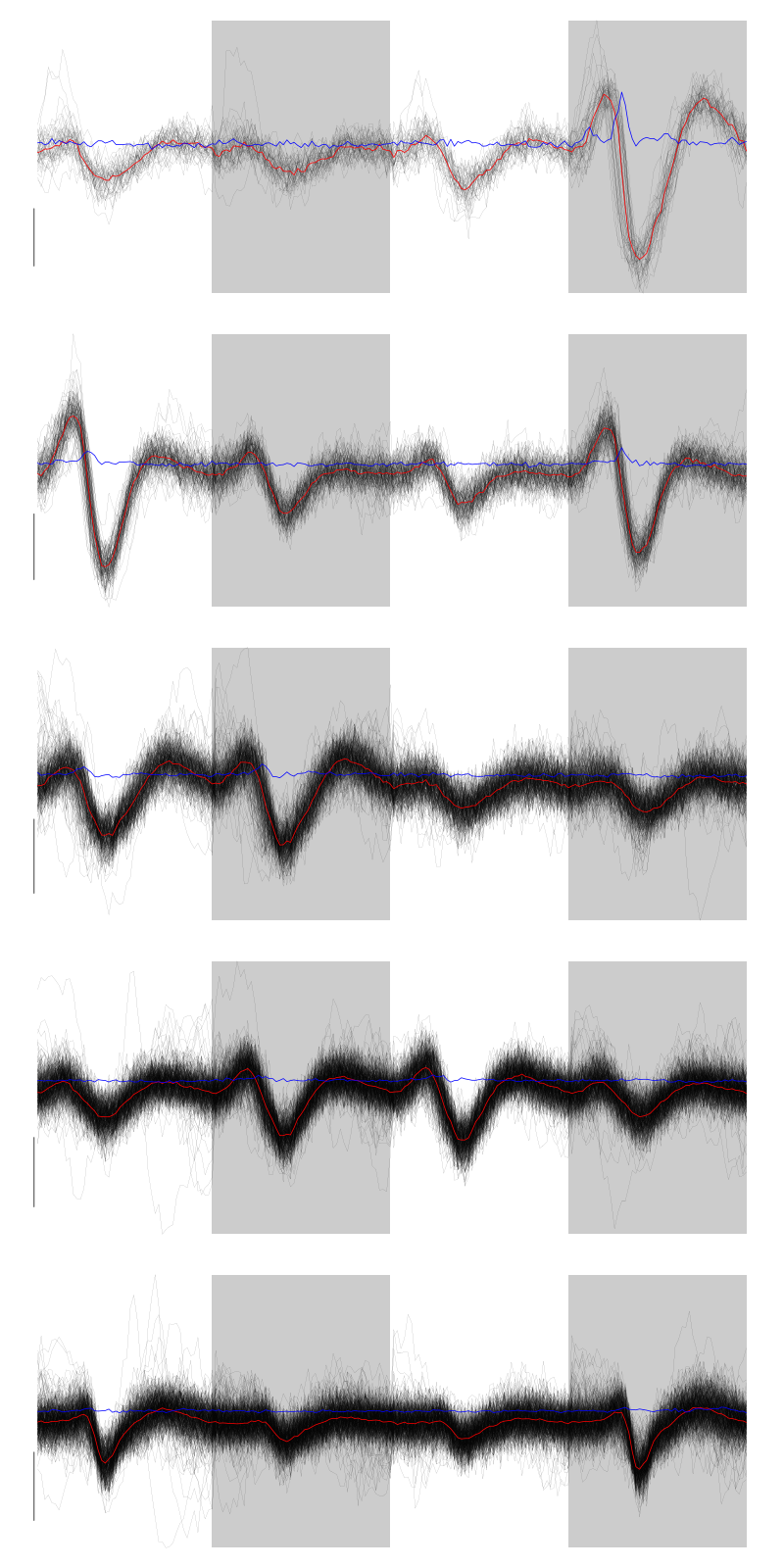
Figure 6: The events of the last five clusters of tetrode B
2.12 Long cuts creation
For the peeling process we need templates that start and end at 0 (we will otherwise generate artifacts when we subtract). We proceed "as usual" with (I tried first with the default value for parameters before and after but I reduced their values after looking at the centers, see the next figure):
c_before = 49
c_after = 80
centers = lapply(1:nbc, function(i)
mk_center_list(sp0[goodEvts][labelb==i],lD,
before=c_before,after=c_after))
names(centers) = paste("Cluster",1:nbc)
We then make sure that our cuts are long enough by looking at them (the first 5):
layout(matrix(1:5,nr=5))
par(mar=c(1,4,1,1))
the_range=c(min(sapply(centers,function(l) min(l$center))),
max(sapply(centers,function(l) max(l$center))))
for (i in 1:5) {
template = centers[[i]]$center
plot(template,lwd=2,col=2,
ylim=the_range,type="l",ylab="")
abline(h=0,col="grey50")
abline(v=(1:2)*(c_before+c_after)+1,col="grey50")
lines(filter(template,rep(1,filter_length)/filter_length),
col=1,lty=3,lwd=2)
abline(h=-threshold_factor,col="grey",lty=2,lwd=2)
lines(centers[[i]]$centerD,lwd=2,col=4)
}
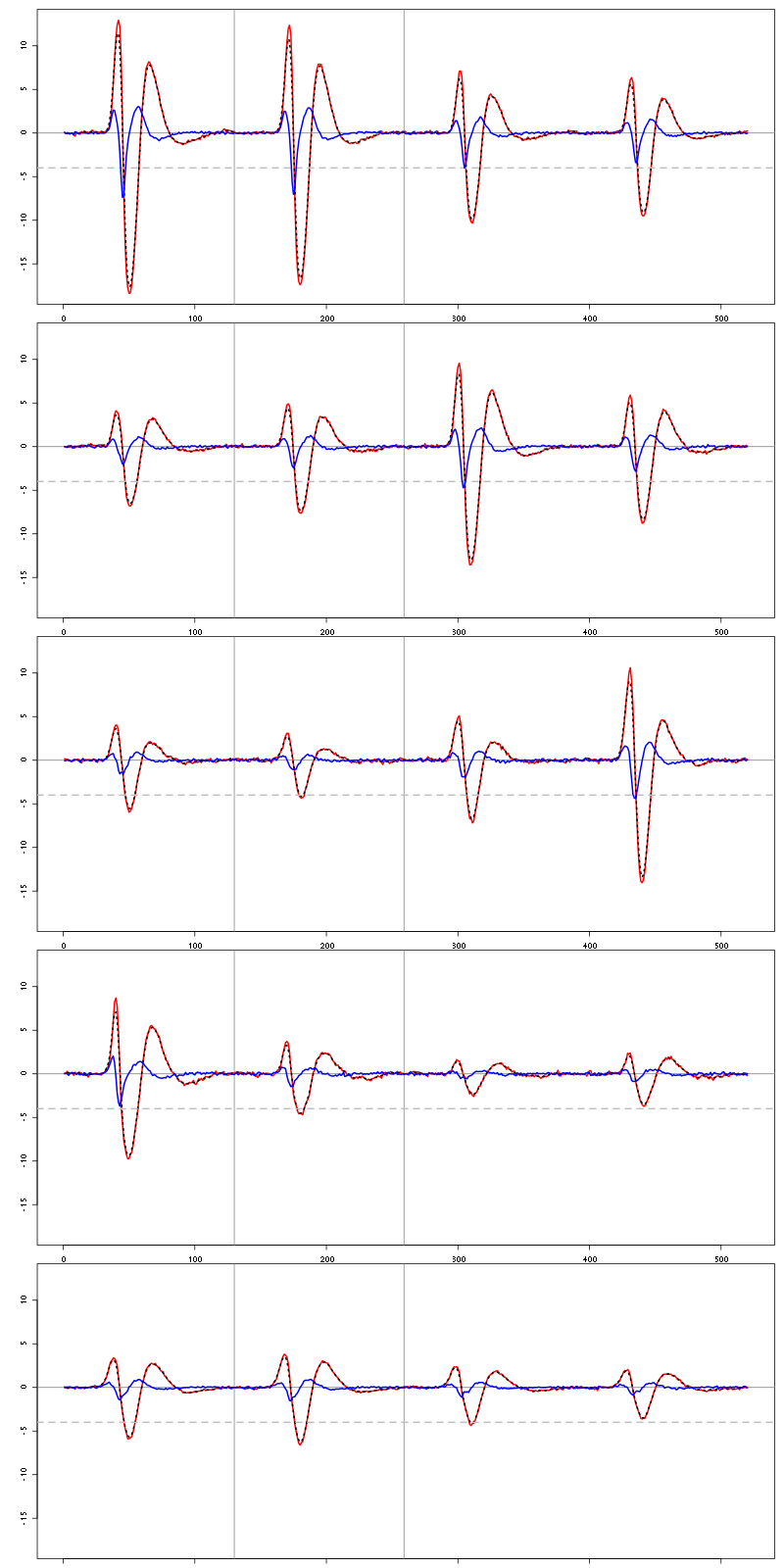
Figure 7: The first five templates (red) together with their first derivative (blue) all with the same scale. The dashed black curve show the templates filtered with the filter used during spike detection and the horizontal dashed grey line shows the detection threshold.
The last five:
layout(matrix(1:5,nr=5))
par(mar=c(1,4,1,1))
the_range=c(min(sapply(centers,function(l) min(l$center))),
max(sapply(centers,function(l) max(l$center))))
for (i in 6:10) {
template = centers[[i]]$center
plot(template,lwd=2,col=2,
ylim=the_range,type="l",ylab="")
abline(h=0,col="grey50")
abline(v=(1:2)*(c_before+c_after)+1,col="grey50")
lines(filter(template,rep(1,filter_length)/filter_length),
col=1,lty=3,lwd=2)
abline(h=-threshold_factor,col="grey",lty=2,lwd=2)
lines(centers[[i]]$centerD,lwd=2,col=4)
}
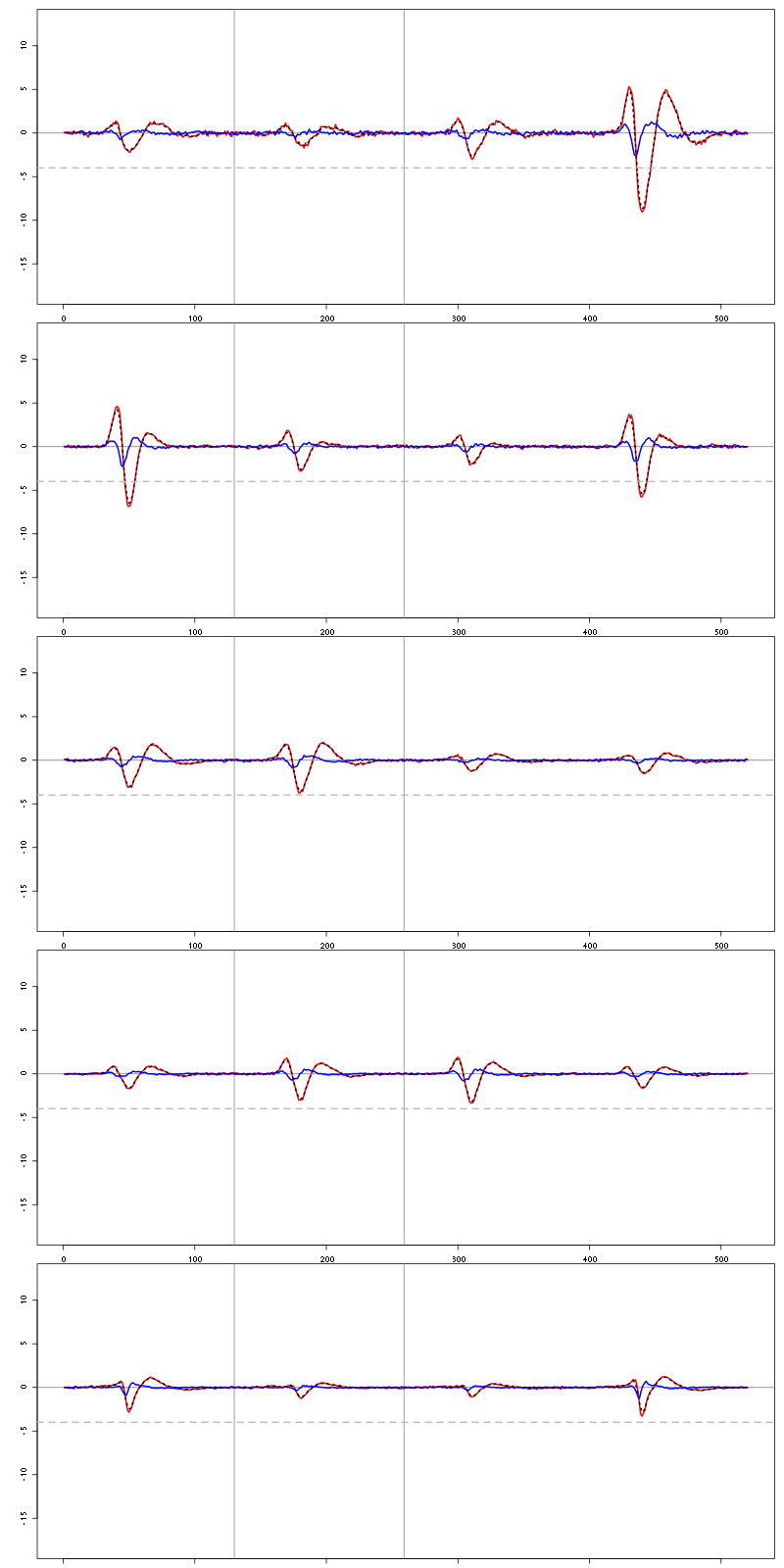

Figure 9: The last five templates (red) together with their first derivative (blue) all with the same scale. The dashed black curve show the templates filtered with the filter used during spike detection and the horizontal dashed grey line shows the detection threshold.
We see that the last three units won't reliably pass our threshold…
2.13 Peeling
We can now do the peeling.
2.13.1 Round 0
We classify, predict, subtract and check how many non-classified events we get:
round0 = lapply(as.vector(sp0),classify_and_align_evt,
data=lD,centers=centers,
before=c_before,after=c_after)
pred0 = predict_data(round0,centers,data_length = dim(lD)[1])
lD_1 = lD - pred0
sum(sapply(round0, function(l) l[[1]] == '?'))
19
We can see the difference before / after peeling for the data between 0.9 and 1.0 s:
ii = 1:1500 + 0.9*15000
tt = ii/15000
par(mar=c(1,1,1,1))
plot(tt, lD[ii,1], axes = FALSE,
type="l",ylim=c(-100,20),
xlab="",ylab="")
lines(tt, lD_1[ii,1], col='red')
lines(tt, lD[ii,2]-30, col='black')
lines(tt, lD_1[ii,2]-30, col='red')
lines(tt, lD[ii,3]-50, col='black')
lines(tt, lD_1[ii,3]-50, col='red')
lines(tt, lD[ii,4]-80, col='black')
lines(tt, lD_1[ii,4]-80, col='red')
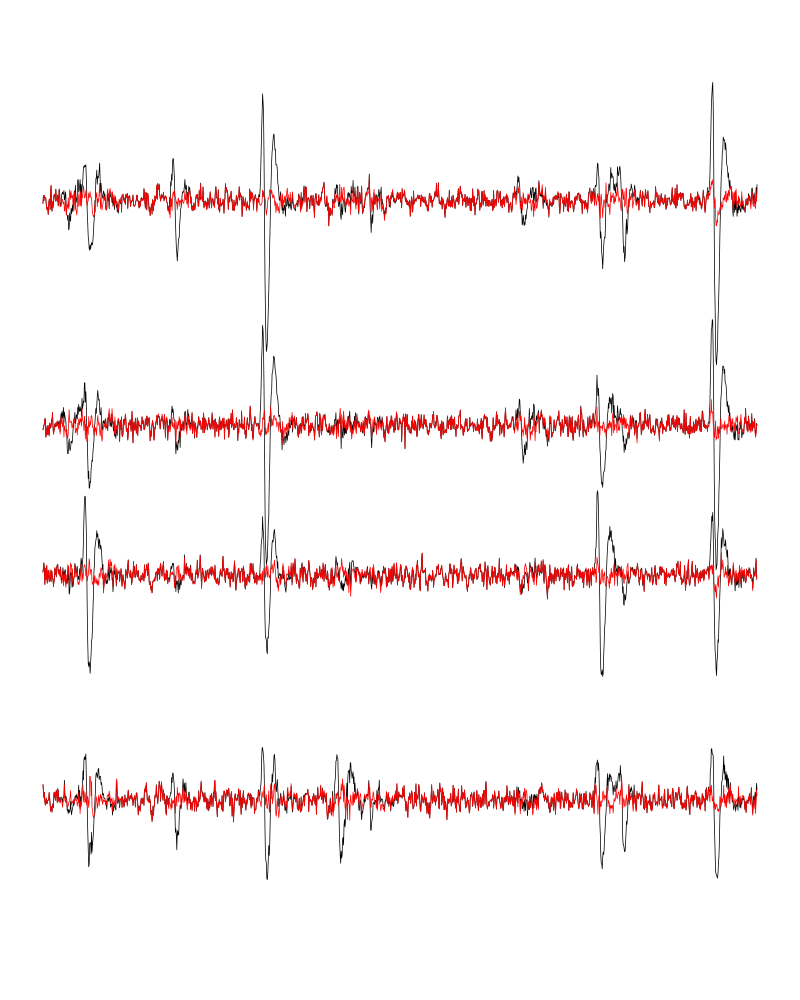
Figure 10: The first peeling illustrated on 100 ms of data, the raw data are in black and the first subtration in red.
2.13.2 Round 1
We keep going, using the subtracted data lD_1 as "raw data", detecting on all sites using the original MAD for normalization:
lDf = -lD_1 lDf = filter(lDf,rep(1,filter_length)/filter_length) lDf[is.na(lDf)] = 0 lDf = t(t(lDf)/lDf_mad_original) thrs = threshold_factor*c(1,1,1,1) bellow.thrs = t(t(lDf) < thrs) lDfr = lDf lDfr[bellow.thrs] = 0 remove(lDf) sp1 = peaks(apply(lDfr,1,sum),15) remove(lDfr) sp1
eventsPos object with indexes of 216 events. Mean inter event interval: 1974.81 sampling points, corresponding SD: 2223.81 sampling points Smallest and largest inter event intervals: 16 and 15982 sampling points.
We classify, predict, subtract and check how many non-classified events we get:
round1 = lapply(as.vector(sp1),classify_and_align_evt,
data=lD_1,centers=centers,
before=c_before,after=c_after)
pred1 = predict_data(round1,centers,data_length = dim(lD)[1])
lD_2 = lD_1 - pred1
sum(sapply(round1, function(l) l[[1]] == '?'))
21
We look at what's left with (not shown):
explore(sp1,lD_2,col=c("black","grey50"))
There is some stuff left but the big guys have disappeared so we decide to stop here.
2.14 Getting the spike trains
round_all = c(round0,round1)
spike_trains = lapply(paste("Cluster",1:nbc),
function(cn) sort(sapply(round_all[sapply(round_all,
function(l) l[[1]]==cn)],
function(l) l[[2]]+l[[3]])))
names(spike_trains) = paste("Cluster",1:nbc)
2.15 Getting the inter spike intervals and the forward and backward recurrence times
2.15.1 ISI distributions
We first get the ISI (inter spike intervals) of each unit:
isi = sapply(spike_trains, diff) names(isi) = names(spike_trains)
We define a plot_isi function that plots the ECDF (empirical cumulative distribution function) of the ISI:
plot_isi = function(isi, ## vector of ISIs
xlab="ISI (s)",
ylab="ECFD",
xlim=c(0,0.5),
sampling_frequency=15000,
... ## additional arguments passed to plot
) {
isi = sort(isi)/sampling_frequency
n = length(isi)
plot(isi,(1:n)/n,type="s",
xlab=xlab,ylab=ylab,
xlim=xlim,...)
}
We get the ISI ECDF for the units with:
layout(matrix(1:(nbc+nbc %% 2),nr=ceiling(nbc/2))) par(mar=c(4,5,6,1)) for (cn in names(isi)) plot_isi(isi[[cn]],main=cn)
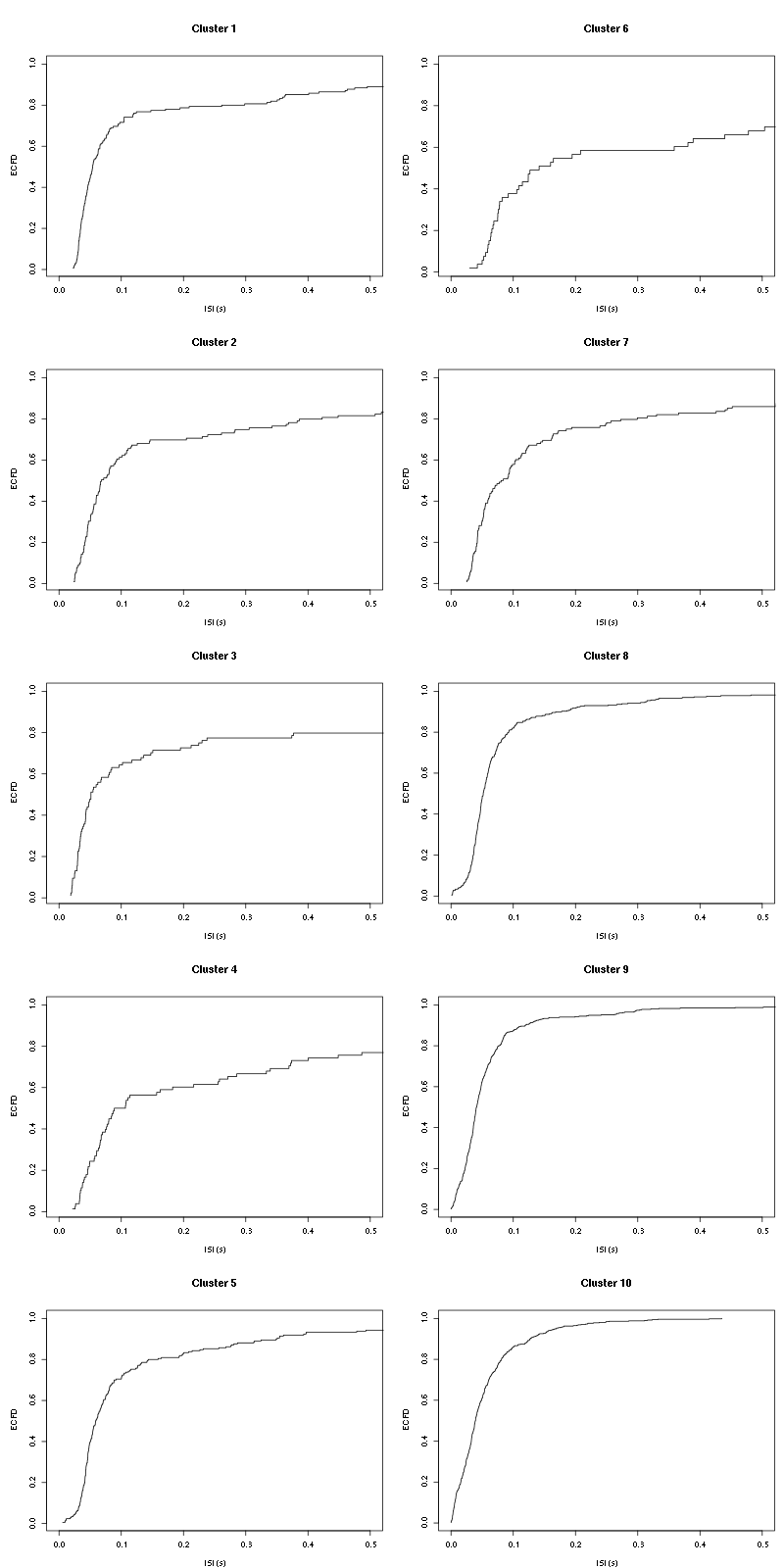
Figure 11: ISI ECDF for the ten units.
The first 7 look great the last three are clearly multi-unit.
2.15.2 Forward and Backward Recurrence Times
The forward recurrence time (FRT) between neuron A and B is the elapsed time between a spike in A and the next spike in B. The backward recurrence time (BRT) is the same thing except that we look for the former spike in B. If A and B are not correlated, the expected density of the FRT is the survival function (1-CDF) of the ISI from B divided by the mean ISI of B (the same holds for the BRT under the null hypothesis after taking the opposite). All that is correct if the data are stationary.
We define a function get_rt that returns the FRT and BRT given two spike trains:
get_rt = function(ref_train,
test_train
) {
res=sapply(ref_train,
function(t) c(max(test_train[test_train<=t])-t,
min(test_train[test_train>=t])-t)
)
res[,!apply(res,2,function(x) any(is.infinite(x)))]
}
We define next a function test_rt that plots a variance stabilized version of the histograms of the FRT and BRT minus the variance stabilized version under the null. The variance stabilization version is replacing the histogram bin counts \(y\) by \(\sqrt{y} + \sqrt{y+1}\). This transformation stabilizes the variance at 1.
test_rt = function(ref_train,
test_train,
sampling_frequency=15000,
nbins=50, ## the number of breaks in the histogram
single_trial_duration = ceiling(max(c(ref_train,test_train))/sampling_frequency),
xlab="Recurrence time (s)",
ylab="Stabilized counts - stabilized expected counts",
subdivisions = 10000, ## argument of integrate
... ## additional parameters passed to plot
) {
rt = ref_train/sampling_frequency
tt = test_train/sampling_frequency
rt_L = vector("list",0)
tt_L = vector("list",0)
idx_max = max(c(rt,tt))%/%single_trial_duration
if ( idx_max == 0) {
rt_L = list(rt)
tt_L = list(tt)
} else {
idx = 0
while (idx <= idx_max) {
start_trial_time = idx*single_trial_duration
end_trial_time = start_trial_time + single_trial_duration
rt_t = rt[start_trial_time <= rt & rt < end_trial_time]
tt_t = tt[start_trial_time <= tt & tt < end_trial_time]
if (length(rt_t) > 0 && length(tt_t) > 0) {
rt_L = c(rt_L,list(rt_t-start_trial_time))
tt_L = c(tt_L,list(tt_t-start_trial_time))
}
idx = idx + 1
}
}
tt_isi_L = lapply(tt_L,diff)
it = unlist(tt_isi_L)
p_it=ecdf(it) ## ECDF of ISI from test
mu_it=mean(it)
s_it=function(t) (1-p_it(t))/mu_it ## expected density of FRT/BRT under the null
## Get the BRT and FRT
res = lapply(1:length(rt_L),
function(idx) {
rt_t = rt_L[[idx]]
tt_t = tt_L[[idx]]
rt_t = rt_t[min(tt_t) < rt_t & rt_t < max(tt_t)]
RT = sapply(rt_t,
function(t) c(max(tt_t[tt_t<=t])-t,
min(tt_t[tt_t>=t])-t)
)
})
frt = sort(unlist(lapply(res, function(l) l[2,])))
brt = sort(-unlist(lapply(res, function(l) l[1,])))
n = length(frt)
frt_h = hist(frt,breaks=nbins,plot=FALSE)
frt_c_s = sqrt(frt_h$counts)+sqrt(frt_h$counts+1) ## stabilized version of the FRT counts
## expected FRT counts under the null
frt_c_e = sapply(1:(length(frt_h$breaks)-1),
function(i) integrate(s_it,frt_h$breaks[i],frt_h$breaks[i+1],subdivisions = subdivisions)$value
)
frt_c_e_s = sqrt(frt_c_e*n) + sqrt(frt_c_e*n+1) ## stabilized version of the expected FRT counts
brt_h = hist(brt,breaks=nbins,plot=FALSE)
brt_c_s = sqrt(brt_h$counts)+sqrt(brt_h$counts+1) ## stabilized version of the BRT counts
## expected BRT counts under the null
brt_c_e = sapply(1:(length(brt_h$breaks)-1),
function(i) integrate(s_it,brt_h$breaks[i],brt_h$breaks[i+1],subdivisions = subdivisions)$value
)
brt_c_e_s = sqrt(brt_c_e*n) + sqrt(brt_c_e*n+1) ## stabilized version of the expected BRT counts
X = c(rev(-brt_h$mids),frt_h$mids)
Y = c(rev(brt_c_s-brt_c_e_s),frt_c_s-frt_c_e_s)
plot(X,Y,type="n",
xlab=xlab,
ylab=ylab,
...)
abline(h=0,col="grey")
abline(v=0,col="grey")
lines(X,Y,type="s")
}
Notice that get_rt is included in test_rt so we don't really need the former anymore.
On the data at hand that gives for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(spike_trains[[i]],
spike_trains[[j]],
ylab="",main=paste("Units",i,"and",j))
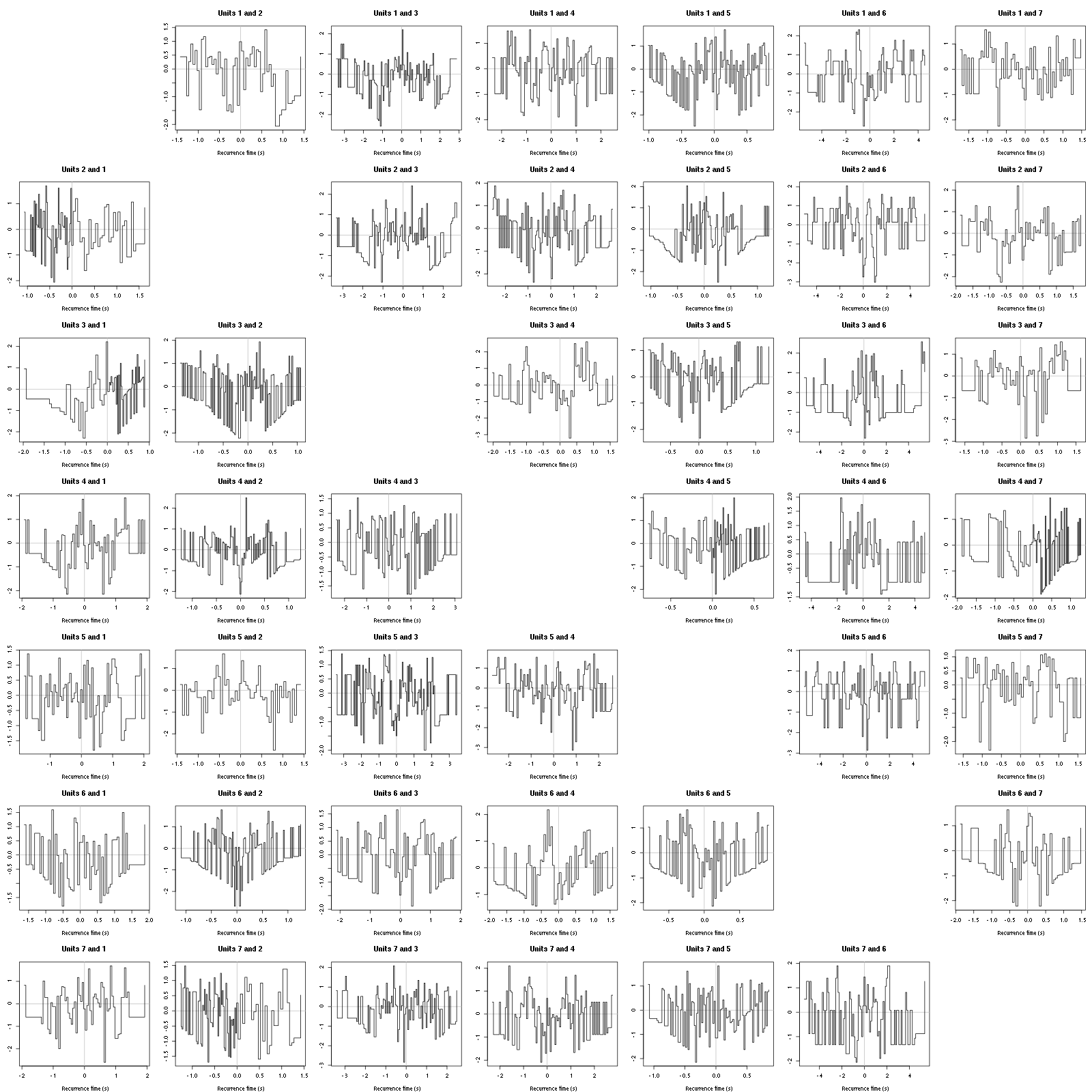
Figure 12: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distrution agaisnt the null hypothesis (no interaction). If the null is correct, the curves should be IID draws from a standard normal distribution.
On this time scale with this number of events, we do not see signs of interactions.
2.16 Making "all at once"
Now that we have classified a single trial we would like to classify the subsequent ones (from the same experiment and the same tetrode) without re-estimating the model every time. If the recording conditions were perfectly stable we could proceed rather easily and in fact to the job in parallel. There are nevertheless few sources of variability we should be ready to cope with:
- First and foremost we can observe a slow drift of the electrodes inducing a change in the relative positions of the neurons and the electrodes. This results in a change of the waveform generated by the neurons on the different recording sites. Since these drifts usually occur on a slow time scale (several minutes) the easiest way to deal with them is to process the data sequentially, in the order in which they were recorded (that precludes the parallel processing alluded to above) and to make the templates evolve slowly by computing, at the end of each trial a weighted average of the templates just used and of the ones obtained by averaging the well isolated events that were just classified.
- A neuron can disappear (because it dies–don't forget that our probes are large and damage the neurons upon insertion–or because of the drift) or appear (because of the drift or "something else" like an hormonal regulation). In the latter case, the model (list of templates) should be updated to include a new element. In any case, the way to detect these appearances / disappearances is to monitor at the end of each trial classification the number of spikes attributed to each neuron of the model as well as the number of unclassified events.
2.16.1 What should our "all at once" function do?
The first thing is to print the five-number summary so that if something went really wrong (amplifier saturation, large noise increase) it will be readily detected. It is then a good idea to print after each peeling round the total number of detected events, the number of events attributed to each neuron and the number of unclassified events. At the end we want to print these same numbers obtained from the whole peeling procedure. The function will also return in a list the following elements:
- prediction: A predicted trace (the overall version of the previous
predX). - residual: A residual trace (the data minus the prediction).
- classification: The
round_allobject above. - unknown: An
eventsobject containing the unclassified events (for quick monitoring in case of problems). - centers: A version of
centerscomputed from the data just analyzed (so that the user can make a weighted average at will). - counts: A vector with the total number of events, the number of events from each neuron and the number of unclassified events.
Argument verbose controls the verbosity: if 0 nothing is printed to the stdout; if 1 the final results are printed; if 2 (default) the final and intermediate (following each peeling round) results are printed. A distinction is made between the first peeling round and the subsequent ones that can have different filter lengths (filter_length_1 and filter_length) and different minimalDist arguments for the peaks function that is called internally. The channels on which the detection is performed at each peeling round is controlled by the corresponding element of vector detection_cycle (a value 0 for an element means that the detection is performed on all channels).
2.16.2 all_at_once definition
So let us define our function:
all_at_once = function(data, ## an unormalized data matrix
centers, ## a list of centers
thres=4*c(1,1), ## threshold vector
filter_length_1=5, ## length of first filter
filter_length=5, ## length of subsequent filters
minimalDist_1=15, ## dead time length imposed at first detection
minimalDist=10, ## dead time length imposed at subsequent detection
before=c_before, ## parameter of centers
after=c_after, ## parameter of centers
detection_cycle=c(0,1,2,3,4), ## where is detection done during the peeling
verbose=2 ## verbosity level
) {
n_samples = dim(data)[1] ## Number of sample points
n_chan = dim(data)[2] ## Number of channels
n_rounds = length(detection_cycle)
if (verbose > 0) {
## print five number summary
cat("The five number summary is:\n")
print(summary(data,digits=2))
cat("\n")
}
## normalize the data
data.mad = apply(data,2,mad)
data = t((t(data)-apply(data,2,median))/data.mad)
filtered_data_mad = sapply(c(filter_length_1,filter_length),
function(l) {
lDf = -data
lDf = filter(lDf,rep(1,l)/l)
lDf[is.na(lDf)] = 0
apply(lDf,2,mad)
})
## Define local function detecting spikes
get_sp = function(dataM,
f_length,
MAD,
site_idx=0,
minimalDist=15) {
lDf = -dataM
lDf = filter(lDf,rep(1,f_length)/f_length)
lDf[is.na(lDf)] = 0
lDf = t(t(lDf)/MAD)
bellow.thrs = t(t(lDf) < thres)
lDfr = lDf
lDfr[bellow.thrs] = 0
if (site_idx == 0)
res = peaks(apply(lDfr,1,sum),minimalDist)
else
res = peaks(lDfr[,site_idx],minimalDist)
res[res>before & res < dim(dataM)[1]-after]
}
out_names = c(names(centers),"?") ## Possible names for classification
data0 = data ## The normalized version of the data
for (r_idx in 1:n_rounds) {
s_idx = detection_cycle[r_idx]
if (verbose > 1 && s_idx==0)
cat(paste("Doing now round",r_idx-1,"detecting on all sites\n"))
if (verbose > 1 && s_idx!=0)
cat(paste("Doing now round",r_idx-1,"detecting on site",s_idx,"\n"))
sp = get_sp(dataM=data,
f_length=ifelse(r_idx==1,filter_length_1,filter_length),
MAD=filtered_data_mad[,ifelse(r_idx==1,1,2)],
site_idx=s_idx,
minimalDist=ifelse(r_idx==1,minimalDist_1,minimalDist))
if (length(sp)==0) next
new_round = lapply(as.vector(sp),classify_and_align_evt,
data=data,centers=centers,
before=before,after=after)
pred = predict_data(new_round,centers,
nb_channels = n_chan,
data_length = n_samples)
data = data - pred
res = sapply(out_names,
function(n) sum(sapply(new_round, function(l) l[[1]] == n)))
res = c(length(sp),res)
names(res) = c("Total",out_names)
if (verbose > 1) {
print(res)
cat("\n")
}
if (r_idx==1)
round_all = new_round
else
round_all = c(round_all,new_round)
}
## Get the global prediction
pred = predict_data(round_all,centers,
nb_channels=n_chan,
data_length=n_samples)
## Get the residuals
resid = data0 - pred
## Repeat inital detection on resid
sp = get_sp(dataM=resid,
f_length=filter_length_1,
MAD=filtered_data_mad[,1],
site_idx=detection_cycle[1],
minimalDist=minimalDist_1)
## make events objects from this stuff
unknown = mkEvents(sp,resid,before,after)
## get global counts
res = sapply(out_names,
function(n) sum(sapply(round_all, function(l) l[[1]] == n)))
res["?"] = length(sp)
res=c(sum(res),res)
names(res) = c("Total",out_names)
if (verbose > 0) {
cat("Global counts at classification's end:\n")
print(res)
}
## Get centers
obs_nb = lapply(out_names[-length(out_names)],
function(cn) sum(sapply(round_all, function(l) l[[1]]==cn)))
names(obs_nb) = out_names[-length(out_names)]
spike_trains = lapply(out_names[-length(out_names)],
function(cn) {
if (obs_nb[cn] <= 1)
return(numeric(0))
else
res = sapply(round_all[sapply(round_all, function(l) l[[1]]==cn)],
function(l)
round(l[[2]]+l[[3]]))
res[res>0 & res<n_samples]
}
)
centersN = lapply(1:length(spike_trains),
function(st_idx) {
if (length(spike_trains[[st_idx]]) == 0)
centers[[st_idx]]
else
mk_center_list(spike_trains[[st_idx]],data0,
before=before,after=after)
}
)
names(centersN) = out_names[-length(out_names)]
list(prediction=pred,
residual=resid,
counts=res,
unknown=unknown,
centers=centersN,
classification=round_all)
}
Argument verbose controls the level of "verbosity": if 0 nothing is printed to the stdout; if 1 the final results are printed; if 2 (default) the final and intermediate (following each peeling round) results are printed.
2.16.3 Test
We test the function with:
## We need again an un-normalized version of the data
ref_data = rbind(cbind(h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch02"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch03"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch05"),
h5read("locust20010214_part1.hdf5", "/Spontaneous_2/trial_30/ch07")))
## We can now use our function
aao=all_at_once(data=ref_data, centers, thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=2)
The five number summary is:
V1 V2 V3 V4
Min. : 98 Min. : 610 Min. : 762 Min. : 475
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3634 Max. :3076 Max. :2887 Max. :3143
Doing now round 0 detecting on all sites
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1950 156 119 84 77 202 48
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
126 285 401 433 19
Doing now round 1 detecting on all sites
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
216 0 1 1 2 9 6
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
3 44 51 78 21
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2185 156 120 85 79 211 54
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
129 329 452 511 59
We see that we are getting back the numbers we obtained before step by step.
We can compare the "old" and "new" centers with (not shown):
layout(matrix(1:nbc,nr=nbc))
par(mar=c(1,4,1,1))
for (i in 1:nbc) {
plot(centers[[i]]$center,lwd=2,col=2,
ylim=the_range,type="l")
abline(h=0,col="grey50")
abline(v=(c_before+c_after)+1,col="grey50")
lines(aao$centers[[i]]$center,lwd=1,col=4)
}
They are not exactly identical since the new version is computed with all events (superposed or not) attributed to each neuron.
3 Analyzing a sequence of trials
3.1 Create a directory were results get saved
We will carry out an analysis of sequences of 30/25 trials with a given odor. At the end of the analysis of the sequence we will save some intermediate R object in a directory we are now creating.:
if (!dir.exists("tetB_analysis"))
dir.create("tetB_analysis")
3.2 Create a data loading function
We want to write a function that takes a trial number and stimulus name and returns a matrix containing stereode Ca (channels 9, 11) data for this trial. The only little trick the function must include is the fact that a number x smaller than 10 appears in the data set name as 0x:
get_tetrode_data = function(trial_idx,stim="Spontaneous_1") {
prefix = ifelse(trial_idx<10,
paste0("/",stim,"/trial_0",trial_idx),
paste0("/",stim,"/trial_",trial_idx)
)
cbind(h5read("locust20010214_part1.hdf5", paste0(prefix,"/ch02")),
h5read("locust20010214_part1.hdf5", paste0(prefix,"/ch03")),
h5read("locust20010214_part1.hdf5", paste0(prefix,"/ch05")),
h5read("locust20010214_part1.hdf5", paste0(prefix,"/ch07")))
}
3.3 sort_many_trials definition
We define now function sort_many_trials. This function takes the following formal parameters:
- inter_trial_time: the time (in sample points) between two successive trials. Example
10*15000. - get_data_fct: a function loading the data of a single trial (like
get_stereode_datawe just defined). This function should take a trial number as its first argument and a sequence name as its second. Exampleget_stereode_data. - stim_name: the sequence name making the second argument of function
get_data_fct. Example "1-Hexanol". - trial_nbs: a vector containing the indices of the trials to sort. Example
1:30. - centers: a list like the
centerslist above. Exampleaao$centers. - counts: a vector with the number of counts each unit got at the previous stage. Example
aao$counts. - all_at_once_call_list: a named list with the arguments used in the previous call to all at once except the first two.
- layout_matrix: a matrix controlling the layout of the diagnostic plots generated while the function is running.
- new_weight_in_update: the maximal weight of the new template in the estimation of the current one.
The function returns a list with the following elements:
- centers: the last estimation of the templates.
- counts: the last counts.
- centers_L: a list of matrices. Each matrix contains the successive template for each neuron (used to track template evolution).
- counts_M: a matrix with the successive counts.
- spike_trains: a list of spike trains, one per neuron.
sort_many_trials = function(inter_trial_time,
get_data_fct,
stim_name,
trial_nbs,
centers,
counts,
all_at_once_call_list=list(thres=4*c(1,1),
filter_length_1=5, filter_length=3,
minimalDist_1=15, minimalDist=10,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(1:10,nr=5),
new_weight_in_update=0.01
) {
centers_old = centers
counts_old = counts
counts_M = matrix(0,nr=length(trial_nbs),nc=length(counts_old))
centers_L = lapply(centers,
function(c) {
res = matrix(0,nr=length(c$center),nc=length(trial_nbs))
res[,1] = c$center
res
}
)
names(centers_L) = names(centers)
nbc = length(centers)
spike_trains = vector("list",nbc)
names(spike_trains) = paste("Cluster",1:nbc)
idx=1
for (trial_idx in trial_nbs) {
ref_data = get_data_fct(trial_idx,stim_name)
cat(paste0("***************\nDoing now trial ",trial_idx," of ",stim_name,"\n"))
analysis = do.call(all_at_once,
c(list(data=ref_data,centers=centers),all_at_once_call_list))
cat(paste0("Trial ",trial_idx," done!\n******************\n"))
centers_new = analysis$centers
counts_new = analysis$counts
counts_M[idx,] = counts_new
centers = centers_new
for (c_idx in 1:length(centers)){
n = counts_new[c_idx+1]
o = counts_old[c_idx+1]
w = new_weight_in_update*ifelse(o>0,min(1,n/o),0) ## New template weight
centers[[c_idx]]$center=w*centers_new[[c_idx]]$center+(1-w)*centers_old[[c_idx]]$center
centers[[c_idx]]$centerD=w*centers_new[[c_idx]]$centerD+(1-w)*centers_old[[c_idx]]$centerD
centers[[c_idx]]$centerDD=w*centers_new[[c_idx]]$centerDD+(1-w)*centers_old[[c_idx]]$centerDD
centers[[c_idx]]$centerD_norm2=sum(centers[[c_idx]]$centerD^2)
centers[[c_idx]]$centerDD_norm2=sum(centers[[c_idx]]$centerDD^2)
centers[[c_idx]]$centerD_dot_centerDD=sum(centers[[c_idx]]$centerD*centers[[c_idx]]$centerDD)
centers_L[[c_idx]][,idx] = centers[[c_idx]]$center
}
layout(layout_matrix)
par(mar=c(1,3,3,1))
the_pch = if (nbc<10) c(paste(1:nbc),"?")
else c(paste(1:9),letters[1:(nbc-9)],"?")
matplot(counts_M[,2:length(counts_old)],type="b",
pch=the_pch)
c_range = range(sapply(centers_L,
function(m) range(m[,1:idx])))
for (i in 1:nbc) {
if (idx<3)
matplot(centers_L[[i]][,1:idx],type="l",col=1,lty=1,lwd=0.5,
main=paste("Unit",i),ylim=c_range)
else
matplot(centers_L[[i]][,1:idx],type="l",col=c(4,rep(1,idx-2),2),
lty=1,lwd=0.5,main=paste("Unit",i),ylim=c_range)
}
centers_old = centers
counts_old = counts_new
round_all = analysis$classification
st = lapply(paste("Cluster",1:nbc),
function(cn) sapply(round_all[sapply(round_all,
function(l) l[[1]]==cn)],
function(l) l[[2]]+l[[3]]))
names(st) = paste("Cluster",1:nbc)
for (cn in paste("Cluster",1:nbc)) {
if (length(st[[cn]]) > 0)
spike_trains[[cn]] = c(spike_trains[[cn]],
(trial_idx-1)*inter_trial_time + sort(st[[cn]]))
}
idx = idx+1
}
spike_trains = lapply(spike_trains,sort)
list(centers=centers,
counts=counts_new,
spike_trains=spike_trains,
counts_M=counts_M,
centers_L=centers_L,
trial_nbs=trial_nbs,
call=match.call())
}
3.4 Diagnostic plot function definitions
We define next three functions defining diagnostic plots at the end of a sequence analysis:
counts_evolution = function(smt_res ## result of a sort_many_trials call
) {
nbc = length(smt_res$centers)
the_pch = if (nbc<10) c(paste(1:nbc),"?")
else c(paste(1:9),letters[1:(nbc-9)],"?")
matplot(smt_res$trial_nbs,
smt_res$counts_M[,2:(nbc+2)],
type="b",pch=the_pch,
main="Counts evolution",xlab="Trial index",ylab="Number of events")
}
waveform_evolution = function(smt_res, ## result of a sort_many_trials call
threshold_factor=4, ## threshold used
layout_matrix=matrix(1:nbc,nr=nbc)
) {
nbc = length(smt_res$centers)
nt = length(smt_res$trial_nbs)
layout(layout_matrix)
par(mar=c(1,3,4,1))
for (i in 1:nbc) {
matplot(smt_res$centers_L[[i]],
type="l",col=c(4,rep(1,nt-2),2),lty=1,lwd=0.5,
main=paste("Unit",i),ylab="")
abline(h=-threshold_factor,col="grey")
}
}
cp_isi=function(smt_res, ## result of a sort_many_trials call
inter_trial_time=10, ## time between trials in seconds
sampling_rate=15000, ## sampling rate in Hz
nbins=50, ## number of bins for isi histogram
isi_max=1, ## largest isi in isi histogram
layout_matrix=matrix(1:(2*still_there),nr=still_there,byrow=TRUE)
) {
t_duration = inter_trial_time
n_trials = length(smt_res$trial_nbs)
nbc = length(smt_res$centers)
still_there = nbc - sum(sapply(smt_res$spike_trains,
function(l)
is.null(l) || length(l) <= n_trials))
layout(layout_matrix)
par(mar=c(4,4,4,1))
for (cn in names(smt_res$spike_trains)) {
if (is.null(smt_res$spike_trains[[cn]]) || length(smt_res$spike_trains[[cn]]) <= n_trials) next
st = smt_res$spike_trains[[cn]]/sampling_rate
plot(st,1:length(st),
main=paste("Observed CP for unit",cn),
xlab="Time (s)",ylab="Nb of evts",type="s")
isi = diff(st)
isi = isi[isi <= isi_max]
hist(isi,breaks=nbins,
prob=TRUE,xlim=c(0,0.5),
main=paste("ISI dist for unit",cn),
xlab="Interval (s)",ylab="Density (1/s)")
}
}
cp_isi_raster=function(smt_res, ## result of a sort_many_trials call
inter_trial_time=10, ## time between trials in seconds
sampling_rate=15000, ## sampling rate in Hz
nbins=50, ## number of bins for isi histogram
isi_max=1, ## largest isi in isi histogram
layout_matrix=matrix(1:(3*still_there),nr=still_there,byrow=TRUE)
) {
t_duration = inter_trial_time
n_trials = length(smt_res$trial_nbs)
nbc = length(smt_res$centers)
still_there = nbc - sum(sapply(smt_res$spike_trains,
function(l)
is.null(l) || length(l) <= n_trials))
layout(layout_matrix)
par(mar=c(4,4,4,1))
for (cn in names(smt_res$spike_trains)) {
if (is.null(smt_res$spike_trains[[cn]]) || length(smt_res$spike_trains[[cn]]) <= n_trials) next
st = smt_res$spike_trains[[cn]]/sampling_rate
plot(st,1:length(st),
main=paste("Observed CP for unit",cn),
xlab="Time (s)",ylab="Nb of evts",type="s")
isi = diff(st)
isi = isi[isi <= isi_max]
hist(isi,breaks=nbins,
prob=TRUE,xlim=c(0,0.5),
main=paste("ISI dist for unit",cn),
xlab="Interval (s)",ylab="Density (1/s)")
plot(c(0,t_duration),c(0,n_trials+1),type="n",axes=FALSE,
xlab="",ylab="",main=paste("Raster of unit",cn))
for (t_idx in 1:n_trials) {
sub_st = st[(t_idx-1)*t_duration <= st &
st < t_idx*t_duration] - (t_idx-1)*t_duration
if (length(sub_st) > 0)
points(sub_st, rep(t_idx,length(sub_st)), pch=".")
}
}
}
4 Systematic analysis of the 30 trials from Spontaneous_2 backwards
We will carry out an analysis of the 30 trials of Spontaenous_2 backwards since the model was estimated from the last trial. The LabBook mentions that that noise is present at trials 23, 24 and 25 so we skip these trials.
4.1 Doing the job
a_Spontaneous_2_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name="Spontaneous_2",
trial_nbs=rev(c(1:22,26:30)),
centers=aao$centers,
counts=aao$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
***************
Doing now trial 30 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 98 Min. : 610 Min. : 762 Min. : 475
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3634 Max. :3076 Max. :2887 Max. :3143
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2193 156 120 85 78 208 53
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
130 337 447 517 62
Trial 30 done!
******************
***************
Doing now trial 29 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 229 Min. : 663 Min. :1029 Min. : 786
1st Qu.:2010 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3339 Max. :3083 Max. :2755 Max. :3033
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2220 118 119 51 143 232 38
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
154 350 349 570 96
Trial 29 done!
******************
***************
Doing now trial 28 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 265 Min. : 652 Min. : 960 Min. : 889
1st Qu.:2011 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3260 Max. :3137 Max. :2837 Max. :2871
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2100 110 164 42 124 284 63
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
137 300 306 493 77
Trial 28 done!
******************
***************
Doing now trial 27 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 412 Min. : 533 Min. :1025 Min. : 742
1st Qu.:2012 1st Qu.:2015 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2094 3rd Qu.:2101
Max. :3304 Max. :3133 Max. :2742 Max. :3102
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2038 120 123 58 60 231 23
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
146 366 368 466 77
Trial 27 done!
******************
***************
Doing now trial 26 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 442 Min. : 715 Min. : 988 Min. : 789
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3263 Max. :3140 Max. :2924 Max. :3014
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2024 92 111 56 123 176 33
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
160 240 414 543 76
Trial 26 done!
******************
***************
Doing now trial 22 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 287 Min. : 529 Min. : 656 Min. : 795
1st Qu.:2012 1st Qu.:2015 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3253 Max. :3077 Max. :3046 Max. :3202
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1942 111 140 59 116 202 23
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
123 289 325 476 78
Trial 22 done!
******************
***************
Doing now trial 21 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 141 Min. : 642 Min. :1081 Min. : 937
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2056 Median :2059
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2094 3rd Qu.:2102
Max. :3474 Max. :3006 Max. :2838 Max. :3148
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1914 128 107 65 122 246 29
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
115 283 282 451 86
Trial 21 done!
******************
***************
Doing now trial 20 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 343 Min. : 645 Min. : 830 Min. : 834
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2056 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2094 3rd Qu.:2102
Max. :3295 Max. :3059 Max. :2915 Max. :2951
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2004 172 112 79 72 232 42
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
151 348 220 475 101
Trial 20 done!
******************
***************
Doing now trial 19 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 527 Min. : 642 Min. : 830 Min. : 796
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2094 3rd Qu.:2102
Max. :3199 Max. :3136 Max. :2914 Max. :3006
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2117 135 138 23 144 193 24
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
180 299 309 562 110
Trial 19 done!
******************
***************
Doing now trial 18 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 449 Min. : 617 Min. : 800 Min. : 825
1st Qu.:2012 1st Qu.:2015 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2102
Max. :3136 Max. :2970 Max. :2887 Max. :2921
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2050 138 145 69 65 215 37
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
134 282 372 519 74
Trial 18 done!
******************
***************
Doing now trial 17 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 270 Min. : 580 Min. :1061 Min. : 598
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2094 3rd Qu.:2101
Max. :3323 Max. :3047 Max. :2830 Max. :3172
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2109 139 134 71 132 204 42
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
116 255 441 479 96
Trial 17 done!
******************
***************
Doing now trial 16 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 397 Min. : 600 Min. : 818 Min. : 684
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3343 Max. :3116 Max. :2995 Max. :3070
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2149 143 136 78 123 236 61
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
153 302 312 512 93
Trial 16 done!
******************
***************
Doing now trial 15 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 392 Min. : 691 Min. : 763 Min. : 736
1st Qu.:2012 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3264 Max. :2989 Max. :2960 Max. :2984
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1901 118 132 49 90 178 30
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
144 280 360 432 88
Trial 15 done!
******************
***************
Doing now trial 14 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 270 Min. : 592 Min. :1018 Min. : 604
1st Qu.:2012 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3213 Max. :3061 Max. :2801 Max. :3016
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1959 135 109 66 69 216 21
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
105 231 441 498 68
Trial 14 done!
******************
***************
Doing now trial 13 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 0 Min. : 0 Min. : 24 Min. : 78
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2058 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2100 3rd Qu.:2095 3rd Qu.:2102
Max. :4073 Max. :4073 Max. :4073 Max. :4072
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2098 110 133 88 66 244 39
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
151 318 382 465 102
Trial 13 done!
******************
***************
Doing now trial 12 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 133 Min. : 487 Min. : 785 Min. : 487
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3344 Max. :2981 Max. :2864 Max. :3228
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2050 141 112 58 62 208 28
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
117 258 335 587 144
Trial 12 done!
******************
***************
Doing now trial 11 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 197 Min. : 522 Min. : 720 Min. : 632
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3292 Max. :3067 Max. :2885 Max. :3102
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1963 114 112 74 85 193 37
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
127 259 303 526 133
Trial 11 done!
******************
***************
Doing now trial 10 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 314 Min. : 715 Min. : 924 Min. : 797
1st Qu.:2011 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3434 Max. :3140 Max. :2998 Max. :3197
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2022 144 152 64 110 222 37
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
150 280 324 438 101
Trial 10 done!
******************
***************
Doing now trial 9 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 332 Min. : 669 Min. :1046 Min. : 721
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3206 Max. :2984 Max. :2869 Max. :3094
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1995 94 152 66 98 208 49
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
145 210 381 462 130
Trial 9 done!
******************
***************
Doing now trial 8 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 103 Min. : 415 Min. : 887 Min. : 572
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3433 Max. :3069 Max. :2863 Max. :3104
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1903 110 111 66 68 177 24
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
141 233 437 433 103
Trial 8 done!
******************
***************
Doing now trial 7 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 151 Min. : 613 Min. : 769 Min. : 670
1st Qu.:2011 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2059 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2102
Max. :3575 Max. :3074 Max. :3080 Max. :3132
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2161 174 138 76 92 272 18
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
157 349 304 458 123
Trial 7 done!
******************
***************
Doing now trial 6 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 263 Min. : 680 Min. : 932 Min. : 472
1st Qu.:2011 1st Qu.:2014 1st Qu.:2016 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2102
Max. :3283 Max. :2953 Max. :2937 Max. :3205
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2091 159 143 90 101 234 18
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
138 301 375 425 107
Trial 6 done!
******************
***************
Doing now trial 5 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 334 Min. : 737 Min. : 956 Min. : 478
1st Qu.:2011 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3238 Max. :2998 Max. :2817 Max. :3235
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1970 167 106 47 99 245 22
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
149 272 322 423 118
Trial 5 done!
******************
***************
Doing now trial 4 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 215 Min. : 471 Min. : 640 Min. : 677
1st Qu.:2011 1st Qu.:2014 1st Qu.:2016 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2102
Max. :3639 Max. :3093 Max. :3016 Max. :3076
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2086 199 166 65 118 237 32
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
126 329 304 384 126
Trial 4 done!
******************
***************
Doing now trial 3 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 340 Min. : 704 Min. :1059 Min. : 418
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3381 Max. :3048 Max. :2805 Max. :3035
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1858 142 127 38 87 211 41
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
154 242 369 360 87
Trial 3 done!
******************
***************
Doing now trial 2 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 121 Min. : 686 Min. :1029 Min. : 627
1st Qu.:2011 1st Qu.:2015 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2104 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2101
Max. :3561 Max. :3051 Max. :2812 Max. :3213
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2009 137 149 57 131 212 49
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
101 281 411 374 107
Trial 2 done!
******************
***************
Doing now trial 1 of Spontaneous_2
The five number summary is:
V1 V2 V3 V4
Min. : 0 Min. : 661 Min. :1099 Min. : 459
1st Qu.:2011 1st Qu.:2014 1st Qu.:2017 1st Qu.:2014
Median :2058 Median :2058 Median :2057 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2105 3rd Qu.:2099 3rd Qu.:2095 3rd Qu.:2102
Max. :3560 Max. :3035 Max. :2869 Max. :3329
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
2122 151 160 44 131 246 42
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
147 311 415 380 95
Trial 1 done!
******************
4.2 Diagnostic plots
The counts evolution is:
counts_evolution(a_Spontaneous_2_tetB)
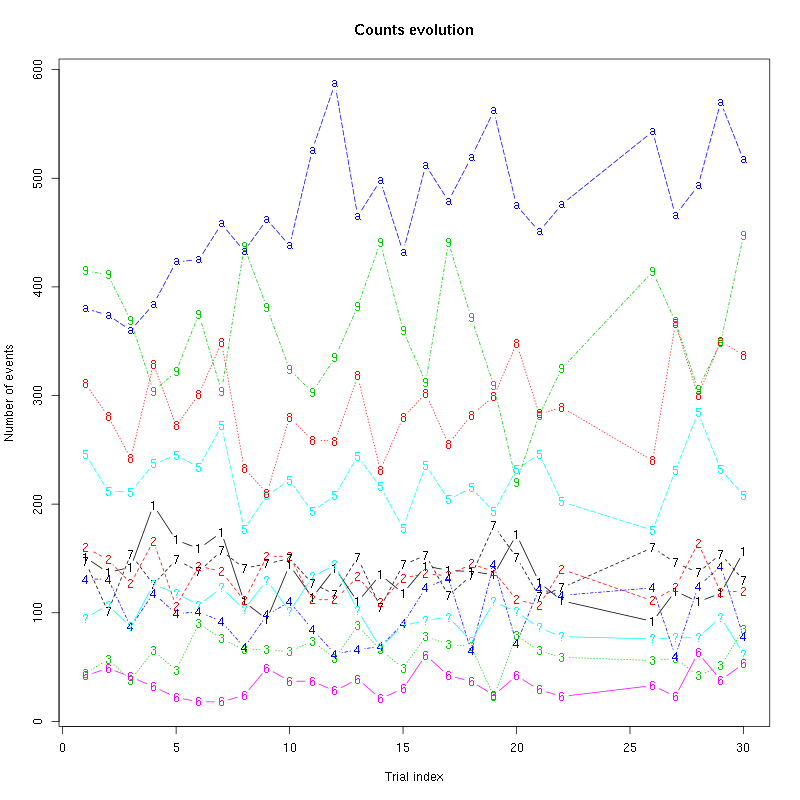
Figure 13: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of Spontaneous_2 for tetrode B.
The waveform evolution is:
waveform_evolution(a_Spontaneous_2_tetB,threshold_factor,matrix(1:10,nr=5))
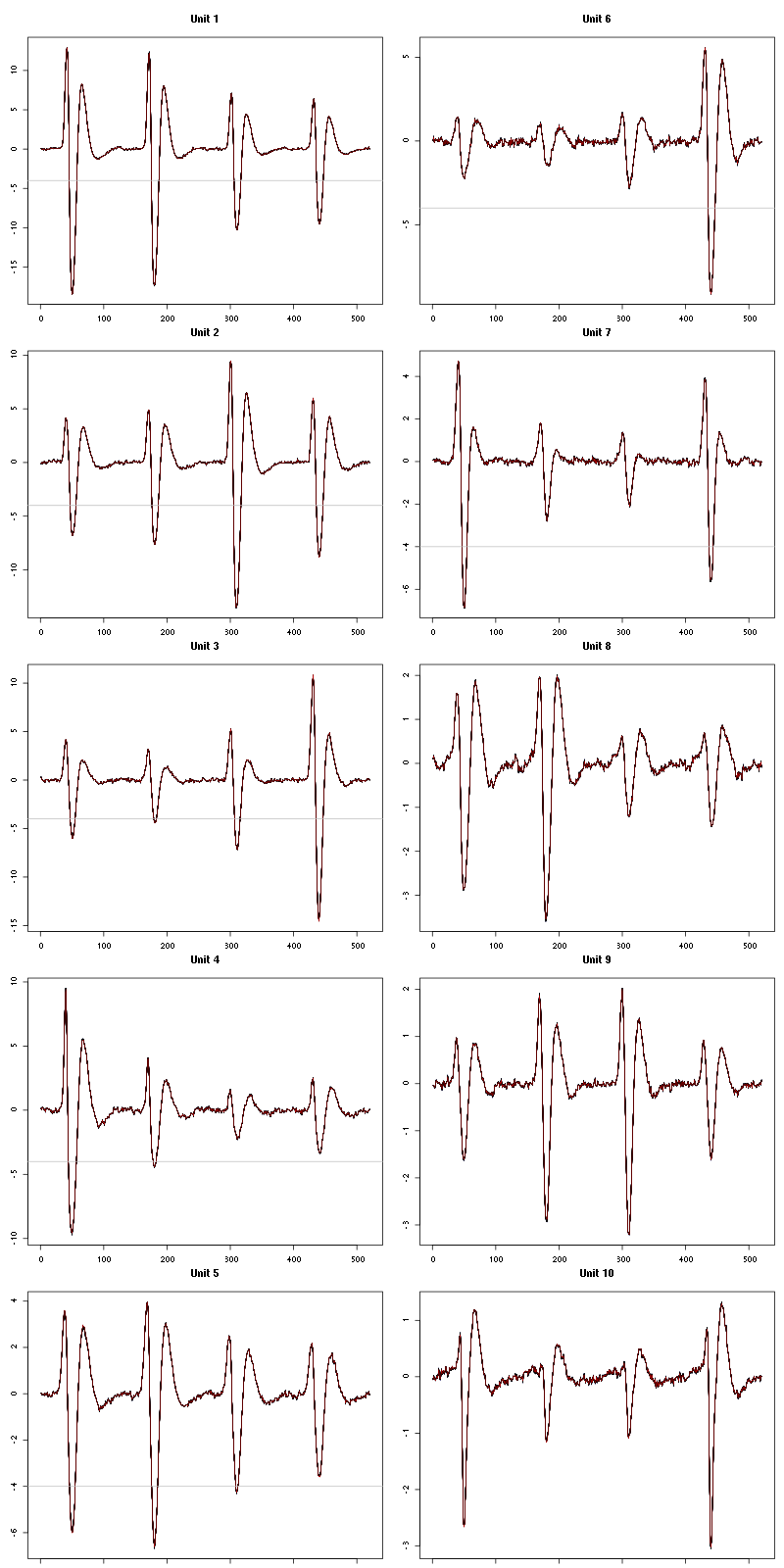
Figure 14: Evolution of the templates of each unit during the 30 trials with Spontaneous_2 for tetrode B.
The observed counting processes and inter spike intervals densities are:
cp_isi(a_Spontaneous_2_tetB,nbins=100)
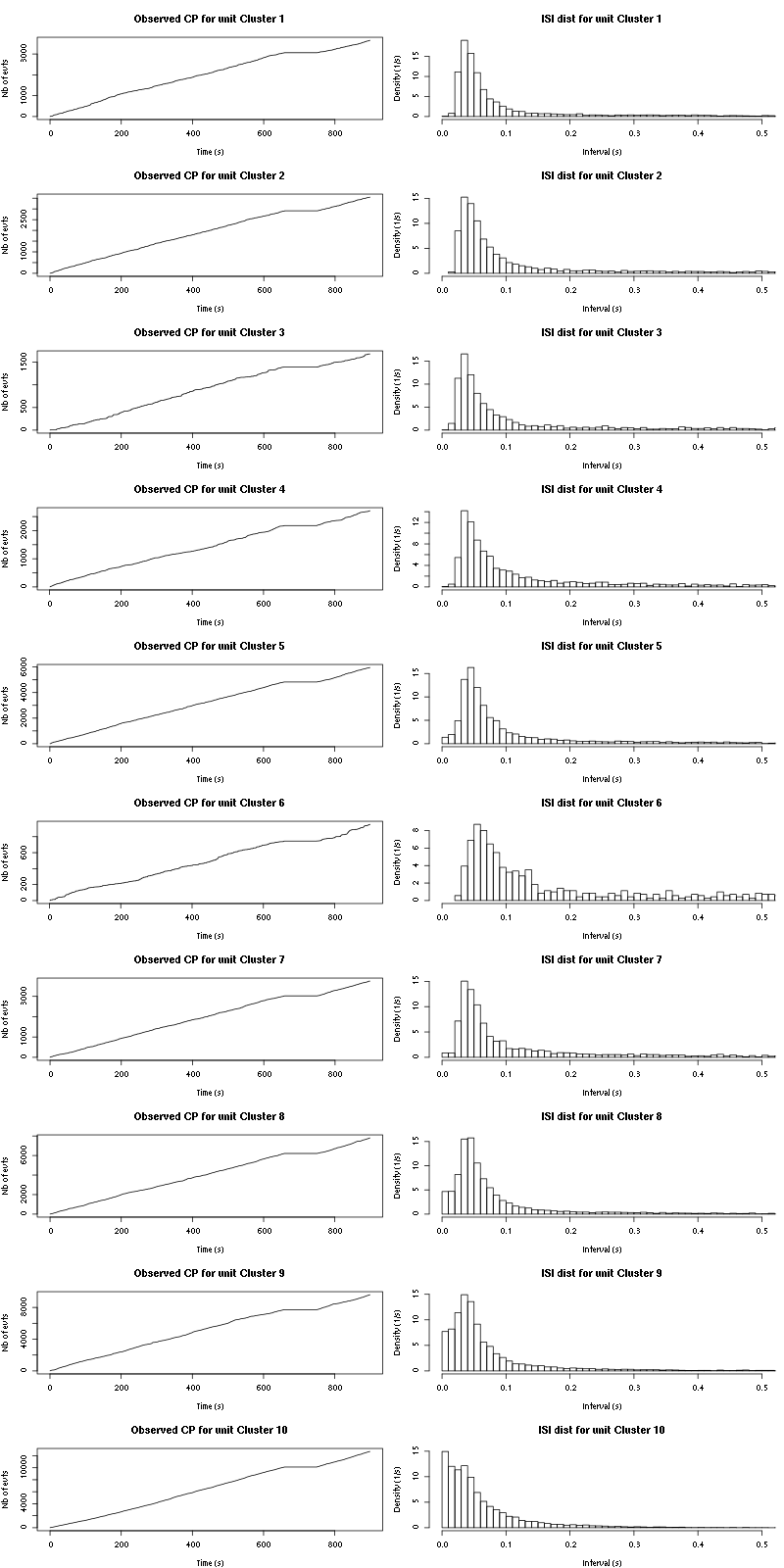
Figure 15: Observed counting processes, empirical inter spike interval distributions and raster plots for Spontaneous_1.
The BRT and RFT tests give for the first 6 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Spontaneous_2_tetB$spike_trains[[i]],
a_Spontaneous_2_tetB$spike_trains[[j]],
nbins=200, single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
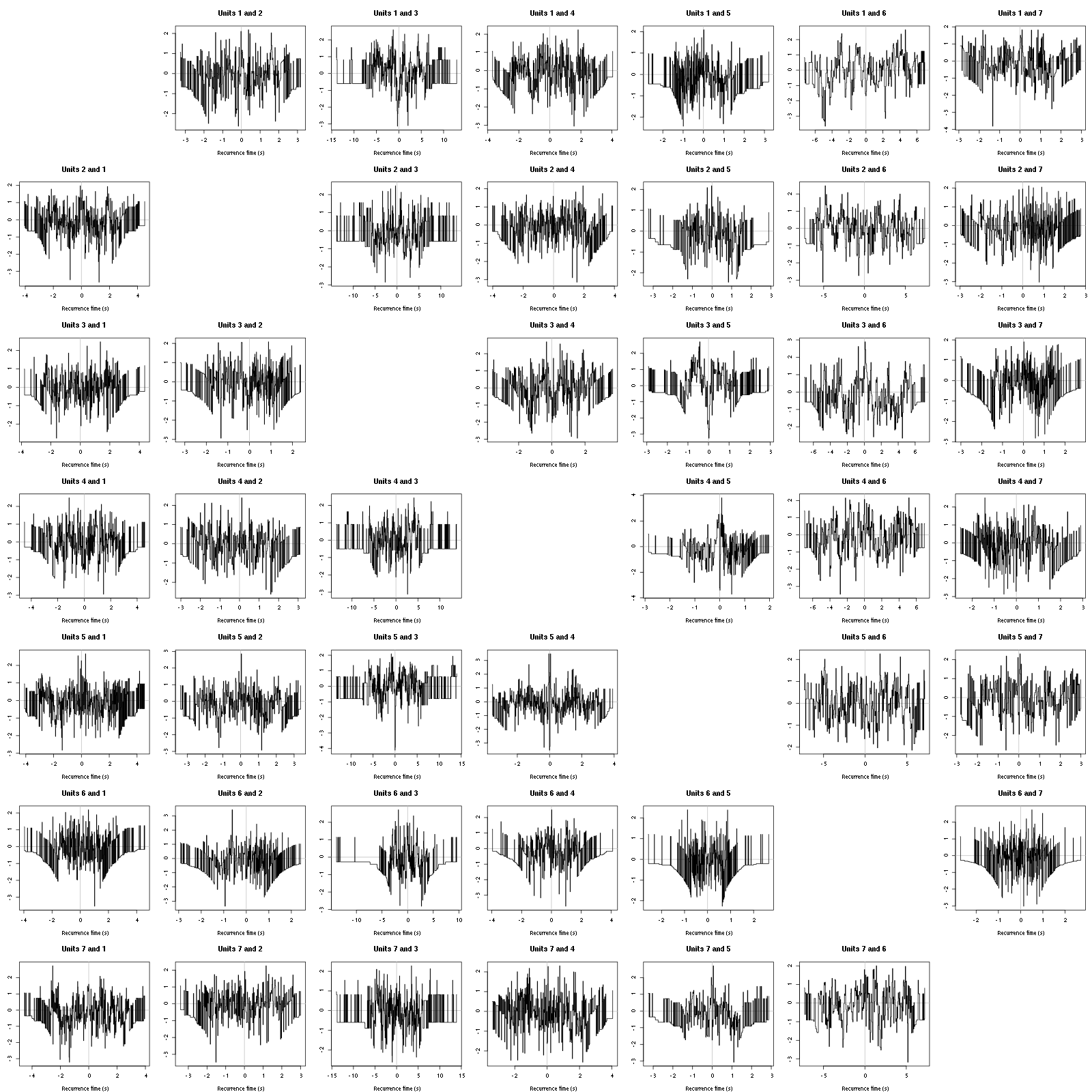
Figure 16: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaneous_2. If the null is correct, the curves should be IID draws from a standard normal distribution.
Still no clear signs of interactions.
4.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Spontaneous_2_tetB,
file=paste0("tetB_analysis/tetB_","Spontaneous_2","_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Spontaneous_2_tetB$spike_trains))
cat(a_Spontaneous_2_tetB$spike_trains[[c_idx]],
file=paste0("locust20010214_spike_trains/locust20010214_Spontaneous_2_tetB_u",c_idx,".txt"),sep="\n")
5 30 trials of Spontaenous_1 backwards
We will carry out an analysis of the 30 trials of Spontaneous_2 backwards. The LabBook mentions that that noise is present at trials 11 and 21 so we skip these trials.
5.1 Do the job
a_Spontaneous_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name="Spontaneous_1",
trial_nbs=rev(c(1:10,12:20,22:30)),
centers=a_Spontaneous_2_tetB$centers,
counts=a_Spontaneous_2_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
***************
Doing now trial 30 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 394 Min. : 741 Min. :1073 Min. : 737
1st Qu.:2013 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3212 Max. :2986 Max. :2984 Max. :3085
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1930 145 147 62 68 186 23
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
113 244 411 401 130
Trial 30 done!
******************
***************
Doing now trial 29 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 343 Min. : 530 Min. : 749 Min. : 821
1st Qu.:2013 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3292 Max. :3109 Max. :2958 Max. :3077
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1790 100 182 61 69 192 32
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
130 214 342 368 100
Trial 29 done!
******************
***************
Doing now trial 28 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 382 Min. : 739 Min. :1006 Min. : 359
1st Qu.:2013 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2102 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2101
Max. :3197 Max. :3089 Max. :2865 Max. :3317
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1819 118 153 52 78 205 31
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
186 239 303 353 101
Trial 28 done!
******************
***************
Doing now trial 27 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 412 Min. : 671 Min. : 888 Min. : 483
1st Qu.:2013 1st Qu.:2016 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2102 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3267 Max. :3011 Max. :2821 Max. :3266
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1718 126 115 58 56 200 46
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
136 238 312 331 100
Trial 27 done!
******************
***************
Doing now trial 26 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 369 Min. : 566 Min. : 900 Min. : 855
1st Qu.:2012 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2059
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2101
Max. :3242 Max. :3101 Max. :2916 Max. :3095
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1923 128 128 61 118 227 55
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
152 260 359 318 117
Trial 26 done!
******************
***************
Doing now trial 25 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 587 Min. : 877 Min. :1091 Min. : 743
1st Qu.:2013 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2103 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2101
Max. :3188 Max. :2939 Max. :2733 Max. :3093
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1767 127 138 57 80 221 27
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
159 243 310 313 92
Trial 25 done!
******************
***************
Doing now trial 24 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 492 Min. : 702 Min. :1130 Min. : 596
1st Qu.:2013 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2102 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3233 Max. :3007 Max. :2723 Max. :3095
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1738 91 110 75 68 182 29
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
171 261 333 337 81
Trial 24 done!
******************
***************
Doing now trial 23 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 362 Min. : 736 Min. :1065 Min. : 430
1st Qu.:2013 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2102 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3241 Max. :3003 Max. :2790 Max. :3203
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1746 103 114 85 75 172 28
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
151 223 409 314 72
Trial 23 done!
******************
***************
Doing now trial 22 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 517 Min. : 779 Min. :1170 Min. : 759
1st Qu.:2013 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2100
Max. :3290 Max. :3095 Max. :2769 Max. :3131
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1744 139 104 34 100 238 29
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
122 276 346 283 73
Trial 22 done!
******************
***************
Doing now trial 20 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 552 Min. : 792 Min. : 970 Min. : 830
1st Qu.:2013 1st Qu.:2016 1st Qu.:2017 1st Qu.:2016
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2094 3rd Qu.:2100
Max. :3186 Max. :2980 Max. :2858 Max. :3093
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1851 133 138 73 47 175 44
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
117 329 390 269 136
Trial 20 done!
******************
***************
Doing now trial 19 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 473 Min. : 656 Min. : 869 Min. : 864
1st Qu.:2013 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3273 Max. :3158 Max. :2926 Max. :3241
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1859 149 140 17 104 275 30
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
120 243 332 284 165
Trial 19 done!
******************
***************
Doing now trial 18 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 518 Min. : 781 Min. :1089 Min. : 377
1st Qu.:2013 1st Qu.:2015 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2098 3rd Qu.:2095 3rd Qu.:2101
Max. :3261 Max. :3000 Max. :2751 Max. :3275
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1835 129 148 38 58 246 57
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
194 255 330 239 141
Trial 18 done!
******************
***************
Doing now trial 17 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 691 Min. : 813 Min. :1043 Min. : 865
1st Qu.:2014 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3154 Max. :2910 Max. :2679 Max. :3093
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1728 129 133 30 48 210 41
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
140 263 356 268 110
Trial 17 done!
******************
***************
Doing now trial 16 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 545 Min. : 990 Min. :1203 Min. : 663
1st Qu.:2014 1st Qu.:2016 1st Qu.:2017 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3104 Max. :2898 Max. :2648 Max. :3121
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1688 113 139 33 59 207 42
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
163 206 366 246 114
Trial 16 done!
******************
***************
Doing now trial 15 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 644 Min. : 986 Min. :1138 Min. : 754
1st Qu.:2013 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2055 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3180 Max. :3002 Max. :2687 Max. :3088
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1724 119 130 39 55 200 25
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
141 237 387 275 116
Trial 15 done!
******************
***************
Doing now trial 14 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 541 Min. : 778 Min. : 975 Min. : 882
1st Qu.:2014 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2057 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3174 Max. :3040 Max. :2734 Max. :3133
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1730 110 130 53 51 180 30
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
140 264 398 269 105
Trial 14 done!
******************
***************
Doing now trial 13 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 531 Min. : 815 Min. :1188 Min. : 797
1st Qu.:2013 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3179 Max. :2946 Max. :2670 Max. :3213
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1760 104 124 33 85 202 35
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
192 222 383 264 116
Trial 13 done!
******************
***************
Doing now trial 12 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 490 Min. : 771 Min. : 975 Min. : 647
1st Qu.:2013 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2102 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2101
Max. :3396 Max. :3005 Max. :2709 Max. :3251
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1867 161 122 45 119 203 27
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
154 270 325 322 119
Trial 12 done!
******************
***************
Doing now trial 10 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 721 Min. : 993 Min. :1250 Min. : 808
1st Qu.:2014 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2093 3rd Qu.:2099
Max. :3103 Max. :2868 Max. :2631 Max. :3095
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1765 88 124 35 34 125 29
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
134 333 341 327 195
Trial 10 done!
******************
***************
Doing now trial 9 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 637 Min. : 921 Min. : 976 Min. : 315
1st Qu.:2014 1st Qu.:2016 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :4071 Max. :4071 Max. :4074 Max. :4074
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1926 154 129 60 50 153 34
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
133 309 303 361 240
Trial 9 done!
******************
***************
Doing now trial 8 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 552 Min. : 789 Min. : 977 Min. : 690
1st Qu.:2014 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2096 3rd Qu.:2094 3rd Qu.:2099
Max. :3106 Max. :2880 Max. :2813 Max. :3236
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1720 113 104 40 68 140 22
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
155 257 319 336 166
Trial 8 done!
******************
***************
Doing now trial 7 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 492 Min. : 957 Min. :1112 Min. : 587
1st Qu.:2014 1st Qu.:2016 1st Qu.:2018 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2100
Max. :3367 Max. :3004 Max. :2744 Max. :3156
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1835 116 145 47 47 158 31
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
149 298 316 345 183
Trial 7 done!
******************
***************
Doing now trial 6 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 672 Min. :1002 Min. :1264 Min. : 763
1st Qu.:2014 1st Qu.:2016 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2096 3rd Qu.:2094 3rd Qu.:2099
Max. :3060 Max. :2879 Max. :2704 Max. :3255
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1810 106 100 52 65 116 37
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
129 282 328 412 183
Trial 6 done!
******************
***************
Doing now trial 5 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 600 Min. : 801 Min. :1080 Min. : 738
1st Qu.:2015 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2093 3rd Qu.:2099
Max. :3162 Max. :2985 Max. :2854 Max. :3103
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1701 107 88 43 64 100 34
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
142 327 323 308 165
Trial 5 done!
******************
***************
Doing now trial 4 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 712 Min. : 971 Min. :1088 Min. : 763
1st Qu.:2014 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2096 3rd Qu.:2094 3rd Qu.:2100
Max. :3196 Max. :2883 Max. :2740 Max. :3245
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1795 128 137 66 65 99 20
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
138 273 376 308 185
Trial 4 done!
******************
***************
Doing now trial 3 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 726 Min. : 985 Min. :1354 Min. : 829
1st Qu.:2014 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2054 Mean :2056
3rd Qu.:2101 3rd Qu.:2096 3rd Qu.:2094 3rd Qu.:2099
Max. :3118 Max. :2873 Max. :2659 Max. :3056
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1801 99 152 38 67 101 24
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
163 303 383 313 158
Trial 3 done!
******************
***************
Doing now trial 2 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 699 Min. : 972 Min. :1209 Min. : 466
1st Qu.:2014 1st Qu.:2017 1st Qu.:2018 1st Qu.:2016
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2056 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2100 3rd Qu.:2096 3rd Qu.:2094 3rd Qu.:2099
Max. :3178 Max. :2922 Max. :2651 Max. :3164
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1718 102 103 48 54 88 44
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
178 287 383 315 116
Trial 2 done!
******************
***************
Doing now trial 1 of Spontaneous_1
The five number summary is:
V1 V2 V3 V4
Min. : 566 Min. : 890 Min. :1158 Min. : 758
1st Qu.:2014 1st Qu.:2016 1st Qu.:2017 1st Qu.:2015
Median :2058 Median :2057 Median :2056 Median :2058
Mean :2055 Mean :2055 Mean :2055 Mean :2056
3rd Qu.:2101 3rd Qu.:2097 3rd Qu.:2094 3rd Qu.:2099
Max. :3159 Max. :2914 Max. :2696 Max. :3179
Global counts at classification's end:
Total Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5 Cluster 6
1807 94 125 32 66 139 31
Cluster 7 Cluster 8 Cluster 9 Cluster 10 ?
181 280 387 350 122
Trial 1 done!
******************
5.2 Diagnostic plots
The counts evolution is:
counts_evolution(a_Spontaneous_1_tetB)
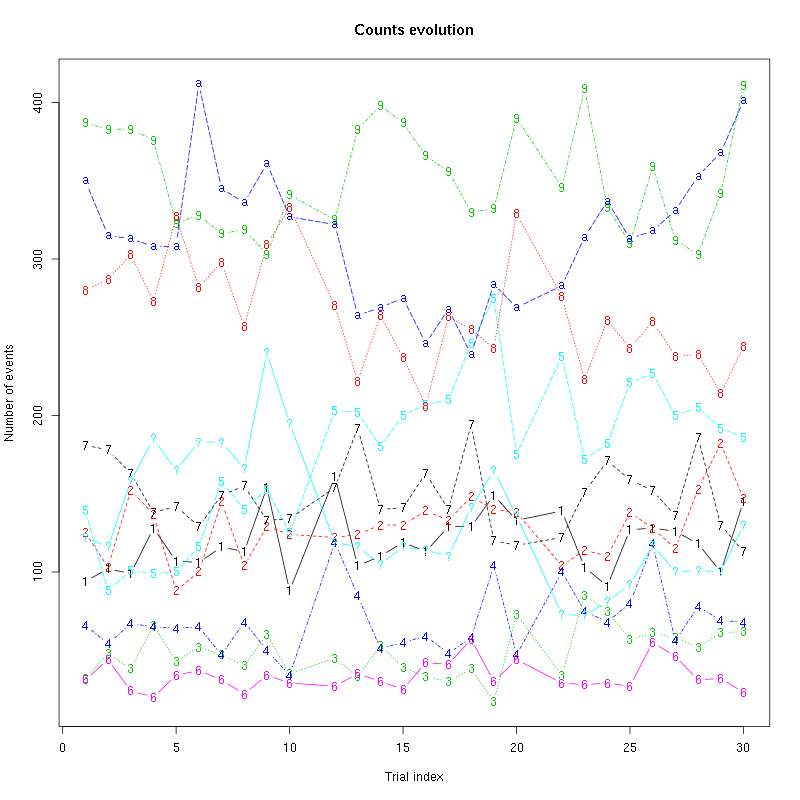
Figure 17: Evolution of the number of events attributed to each unit (1 to 9and "a") or unclassified ("?") during the 30 trials of Spontaneous_1 for tetrode B.
The waveform evolution is:
waveform_evolution(a_Spontaneous_1_tetB,threshold_factor,matrix(1:10,nr=5))
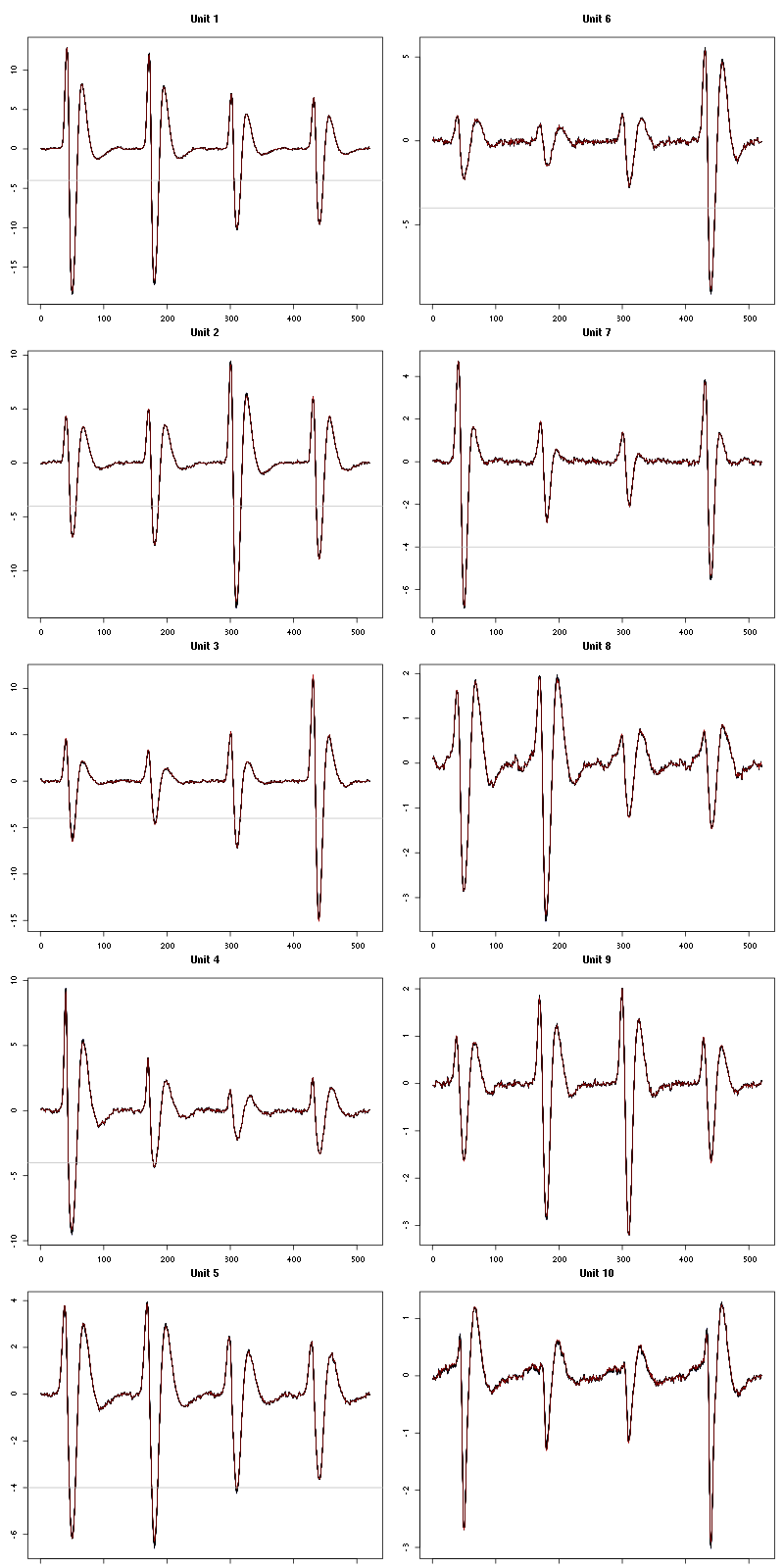
Figure 18: Evolution of the templates of each unit during the 30-2 trials of Spontaneous_1 for tetrode B.
The observed counting processes and inter spike intervals densities are:
cp_isi(a_Spontaneous_1_tetB,nbins=100)
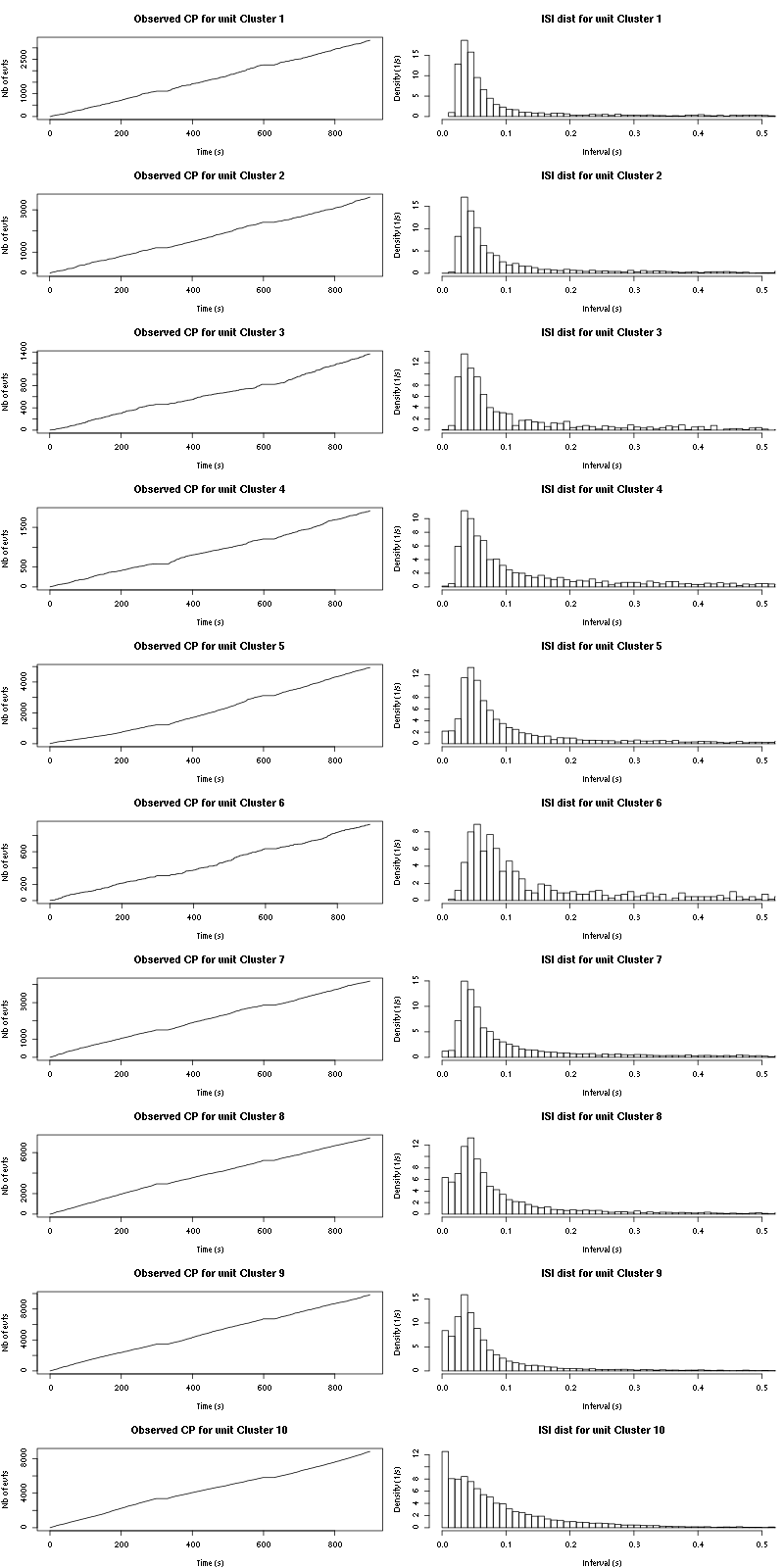
Figure 19: Observed counting processes and empirical inter spike interval distributions for Spontaneous_1.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Spontaneous_1_tetB$spike_trains[[i]],
a_Spontaneous_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
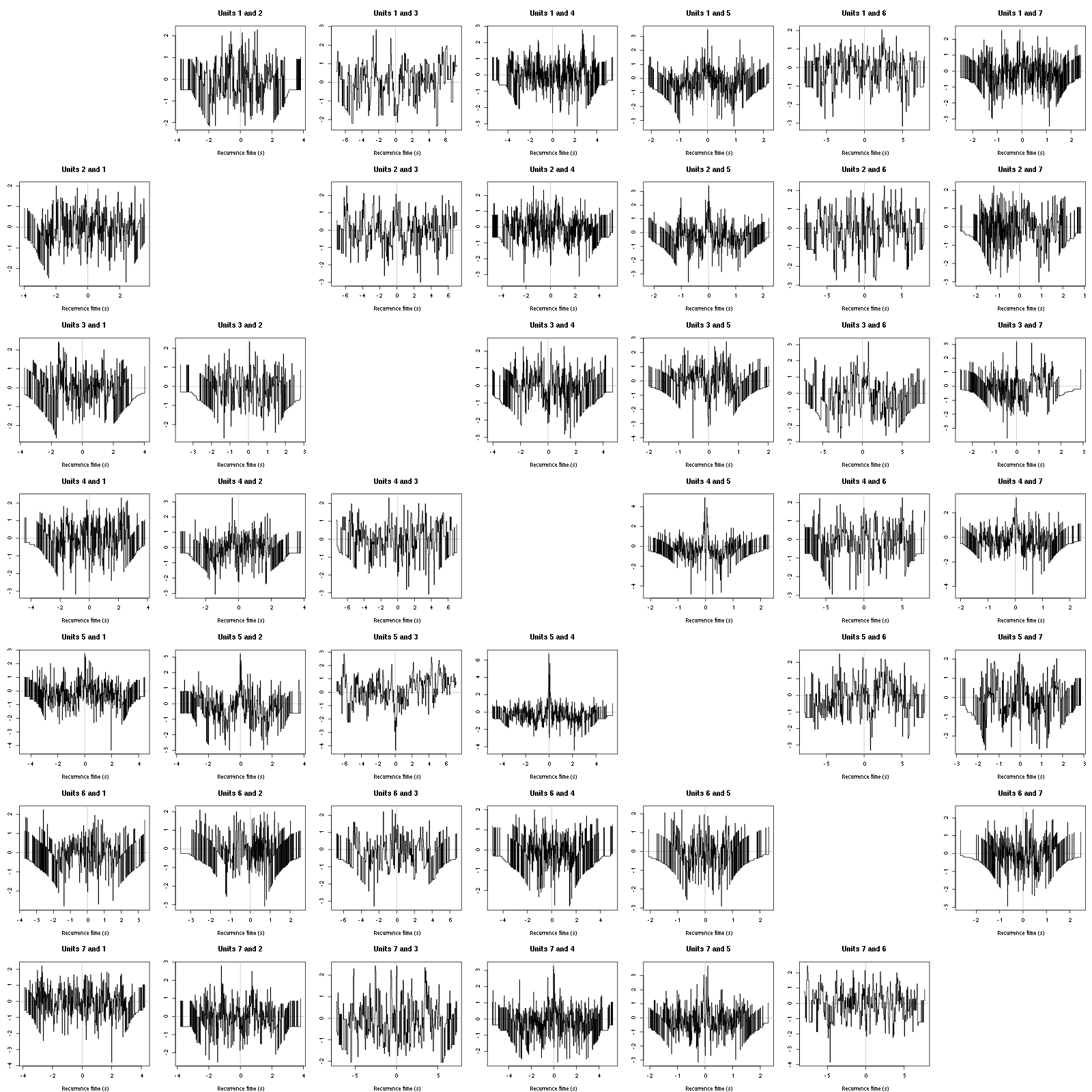
Figure 20: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
5.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Spontaneous_1_tetB,
file=paste0("tetB_analysis/tetB_","Spontaneous_1","_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Spontaneous_1_tetB$spike_trains))
cat(a_Spontaneous_1_tetB$spike_trains[[c_idx]],
file=paste0("locust20010214_spike_trains/locust20010214_Spontaneous_1_tetB_u",c_idx,".txt"),sep="\n")
6 25 trials with Cis-3-Hexen-1-ol (C3H_1)
We will carry out an analysis of the 25 trials from C3H_1.
6.1 Do the job
We do not print out the output to save space.
a_C3H_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name="C3H_1",
trial_nbs=1:25,
centers=aao$centers,
counts=aao$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
6.2 Diagnostic plots
The counts evolution is:
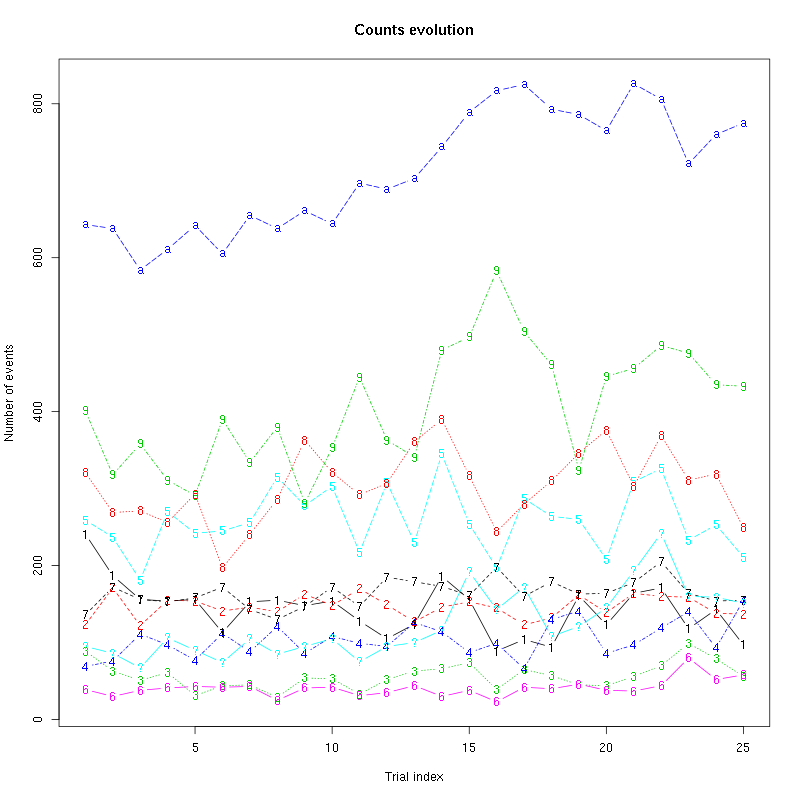
Figure 21: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of C3H_1 for tetrode B.
The waveform evolution is:
waveform_evolution(a_C3H_1_tetB,threshold_factor,matrix(1:10,nr=5))
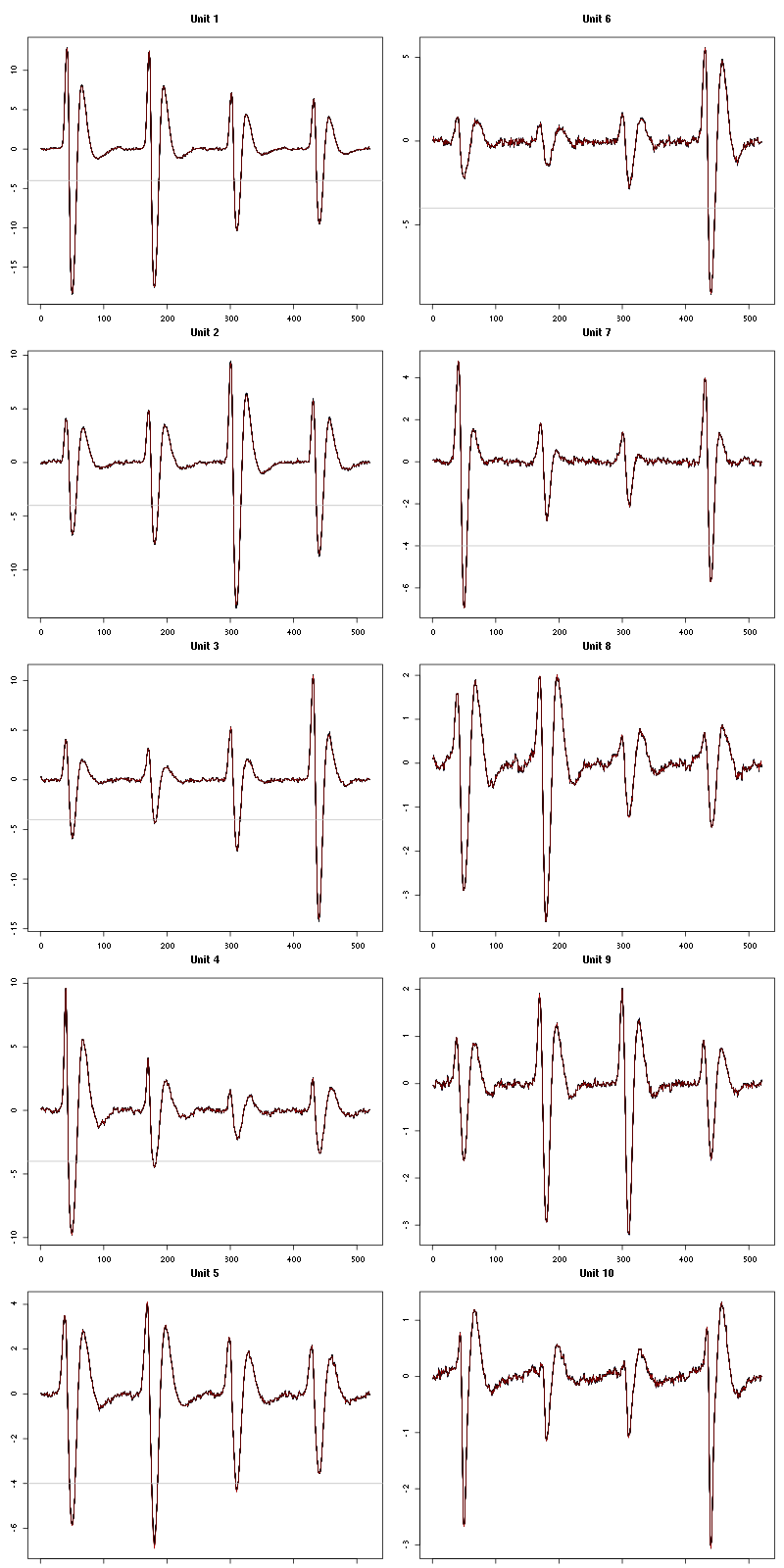
Figure 22: Evolution of the templates of each unit during the 25 trials of C3H_1 for stereode Ca.
The observed counting processes, inter spike intervals densities and raster plots are:
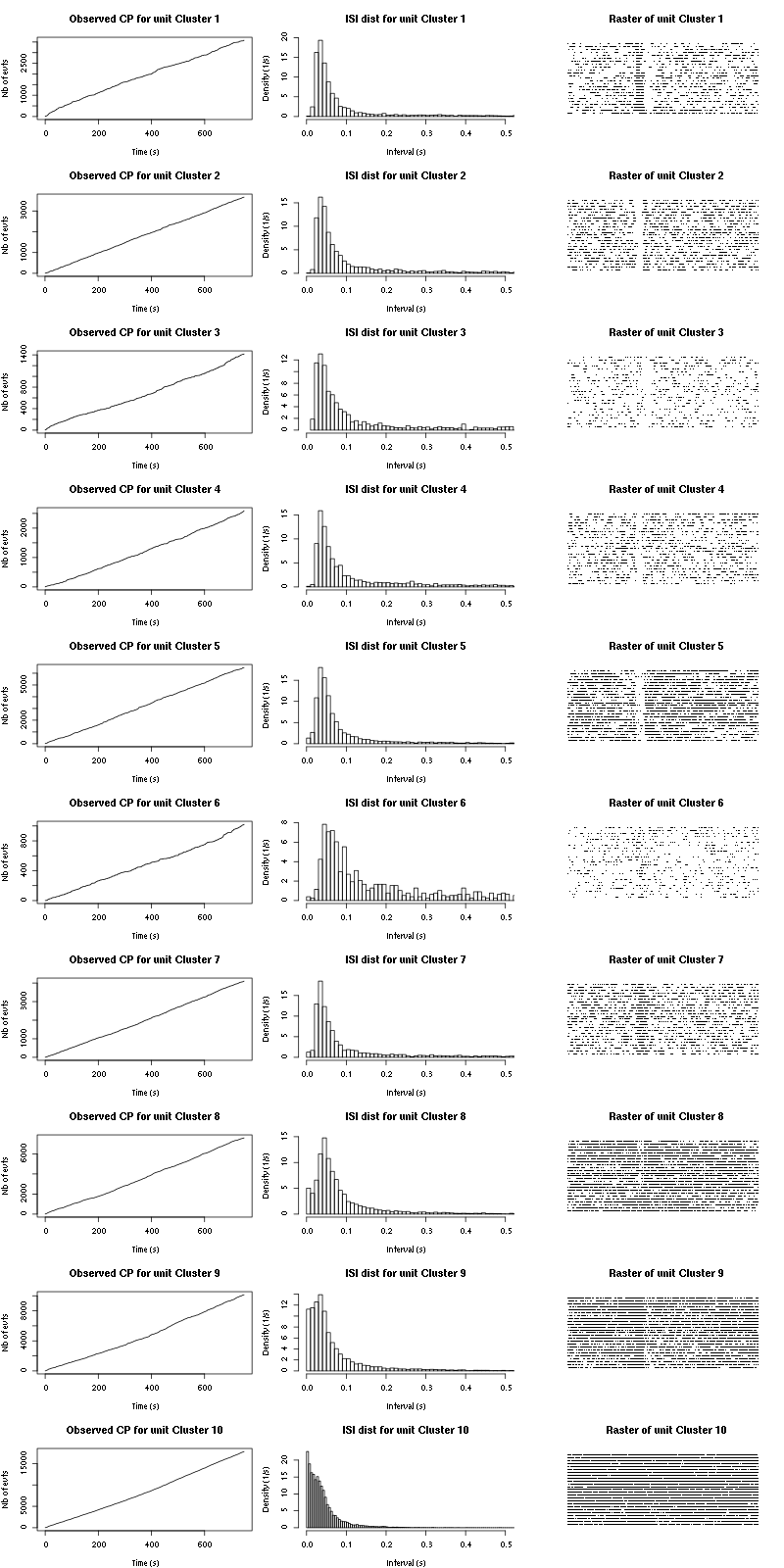
Figure 23: Observed counting processes, empirical inter spike interval distributions and raster plots for C3H_1.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_1_tetB$spike_trains[[i]],
a_C3H_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
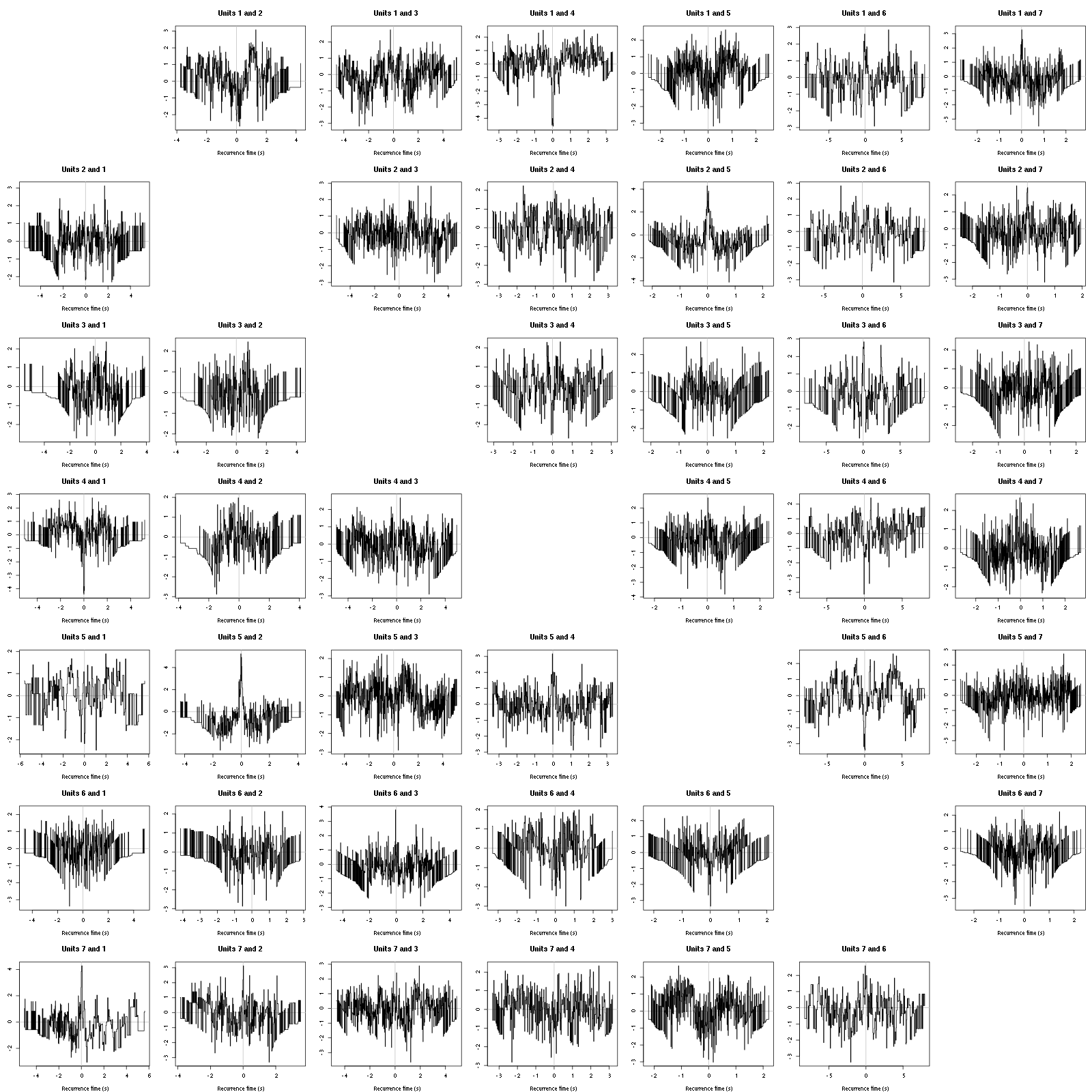
Figure 24: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
6.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_1_tetB,
file=paste0("tetB_analysis/tetB_","C3H_1","_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_1_tetB$spike_trains))
if (!is.null(a_C3H_1_tetB$spike_trains[[c_idx]]))
cat(a_C3H_1_tetB$spike_trains[[c_idx]],
file=paste0("locust20010214_spike_trains/locust20010214_C3H_1_tetB_u",c_idx,".txt"),sep="\n")
7 25 trials with Citral
We will carry out an analysis of the 25 trials from Citral_1.
7.1 Do the job
stim_name = "Citral_1"
a_Citral_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:25,
centers=a_C3H_1_tetB$centers,
counts=a_C3H_1_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
7.2 Diagnostic plots
The counts evolution is:
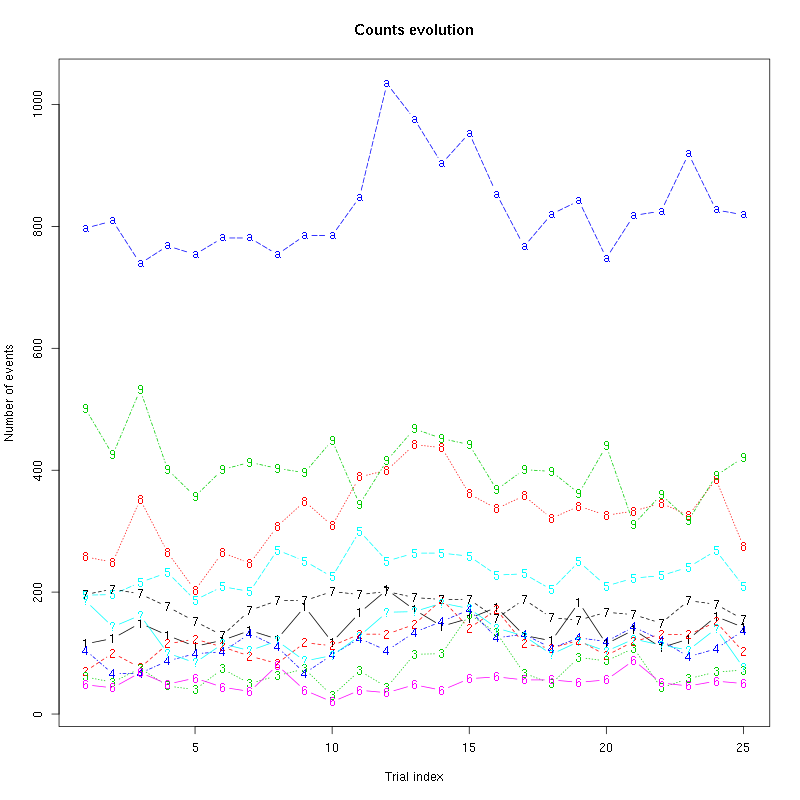
Figure 25: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 25 trials of Citral_1 for tetrodeB.
The waveform evolution is:
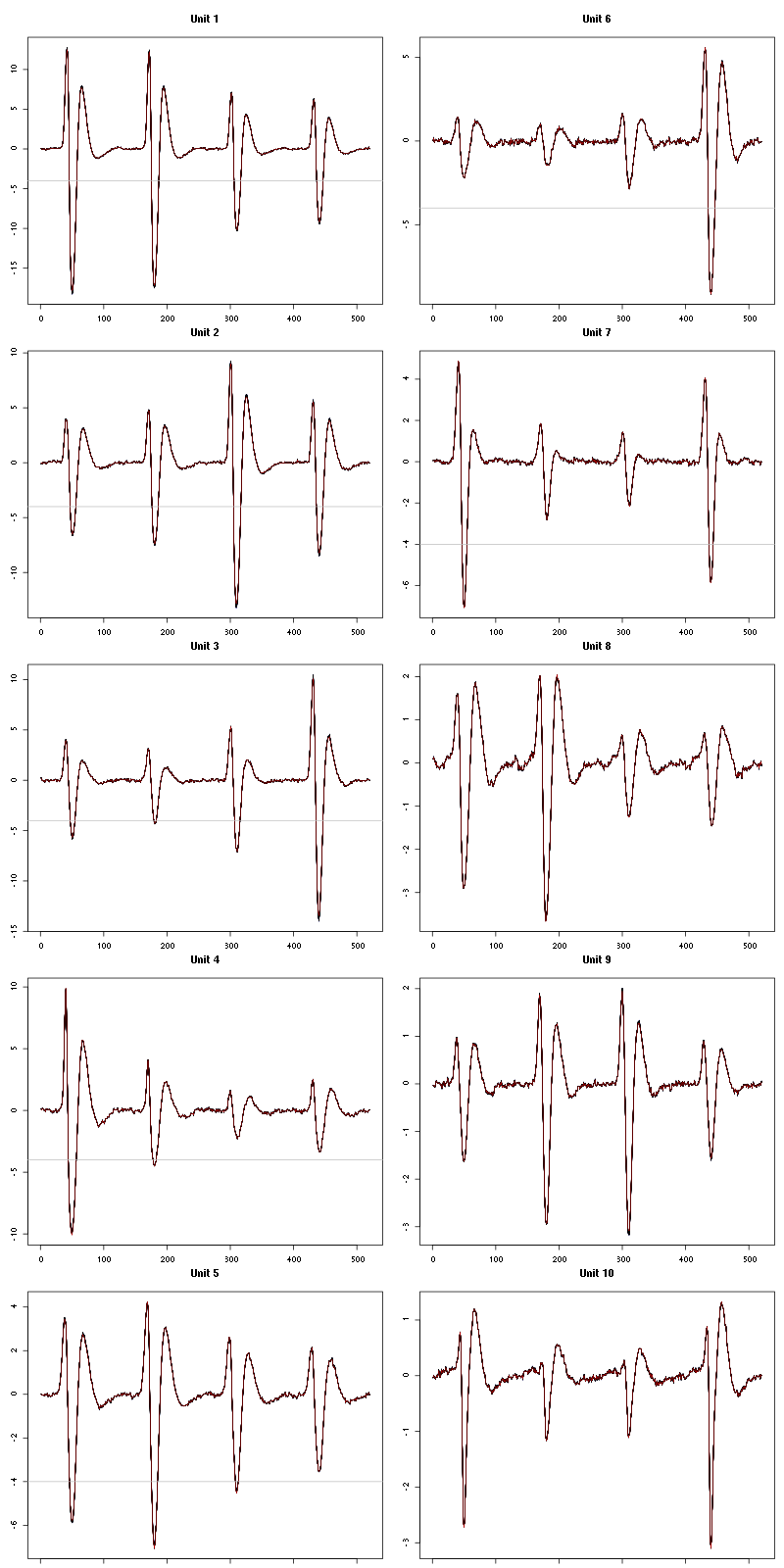
Figure 26: Evolution of the templates of each unit during the 25 trials of Citral for tetrodeB.
The observed counting processes, inter spike intervals densities and raster plots are:
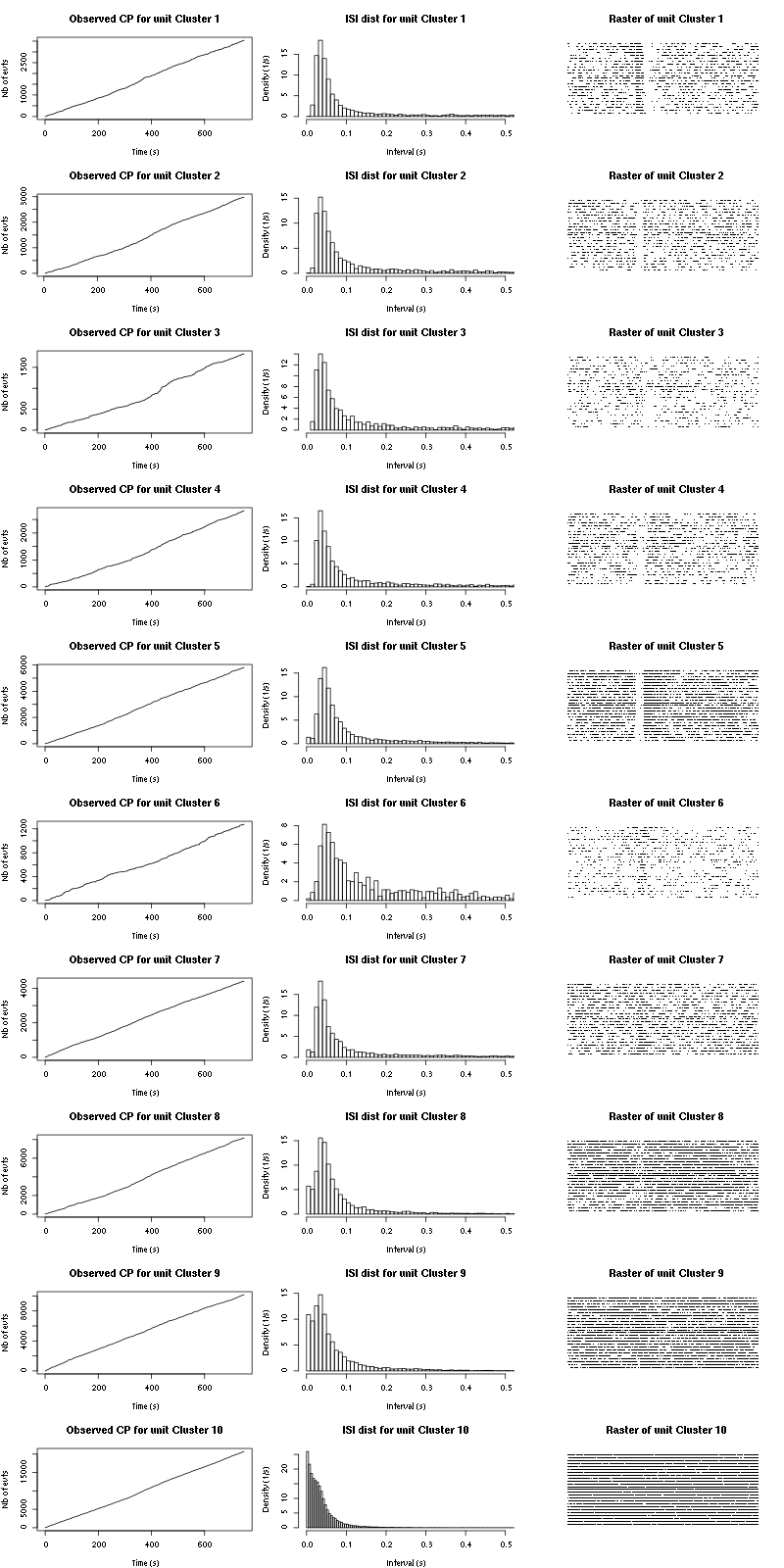
Figure 27: Observed counting processes, empirical inter spike interval distributions and raster plots for Citral.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Citral_1_tetB$spike_trains[[i]],
a_Citral_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
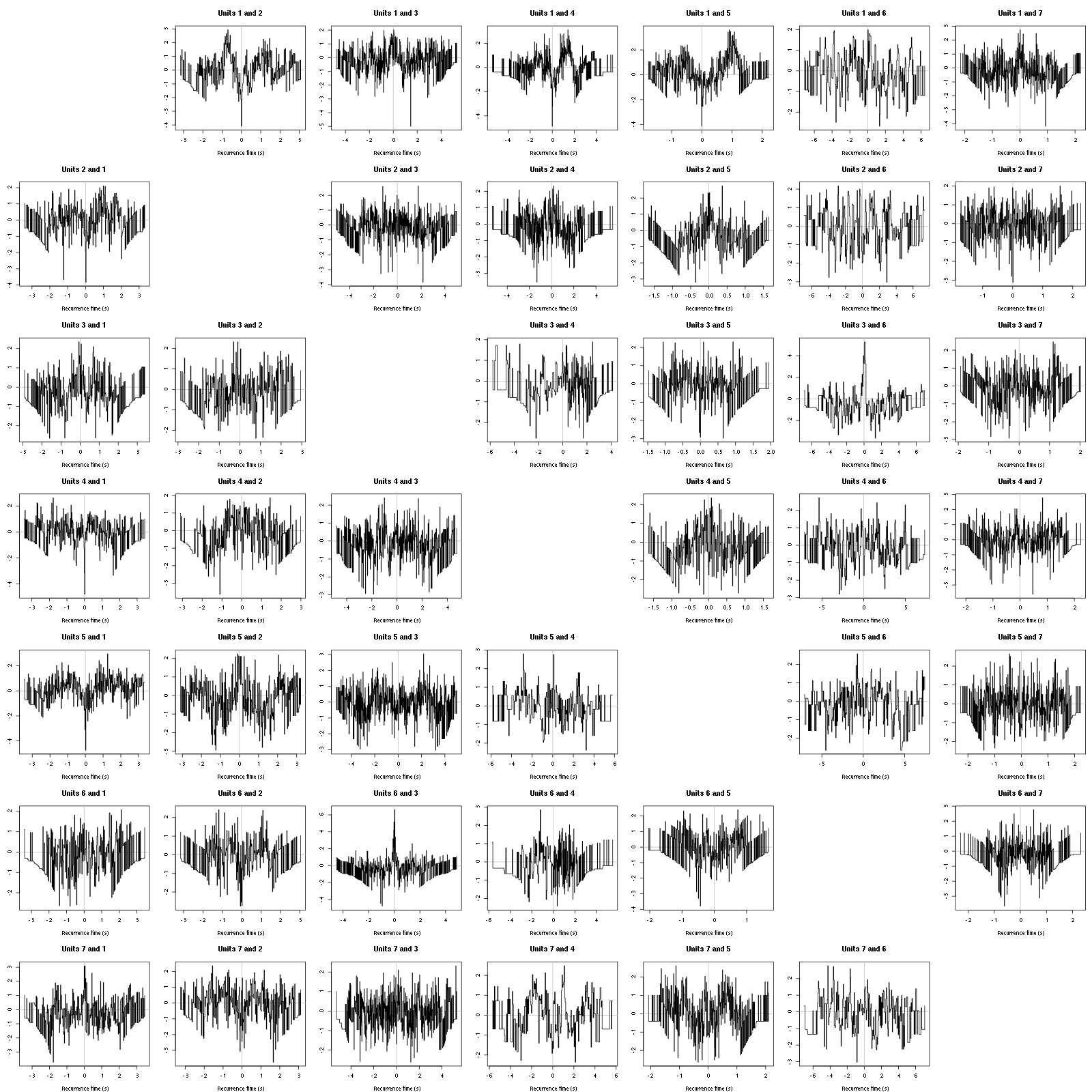
Figure 28: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
7.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Citral_1_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Citral_1_tetB$spike_trains))
if (!is.null(a_Citral_1_tetB$spike_trains[[c_idx]]))
cat(a_Citral_1_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Citral_tetB_u",c_idx,".txt"),sep="\n")
8 25 trials with Vanilla
We will carry out an analysis of the 25 trials from Vanilla_1.
8.1 Do the job
stim_name = "Vanilla_1"
a_Vanilla_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:25,
centers=a_Citral_1_tetB$centers,
counts=a_Citral_1_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
8.2 Diagnostic plots
The counts evolution is:
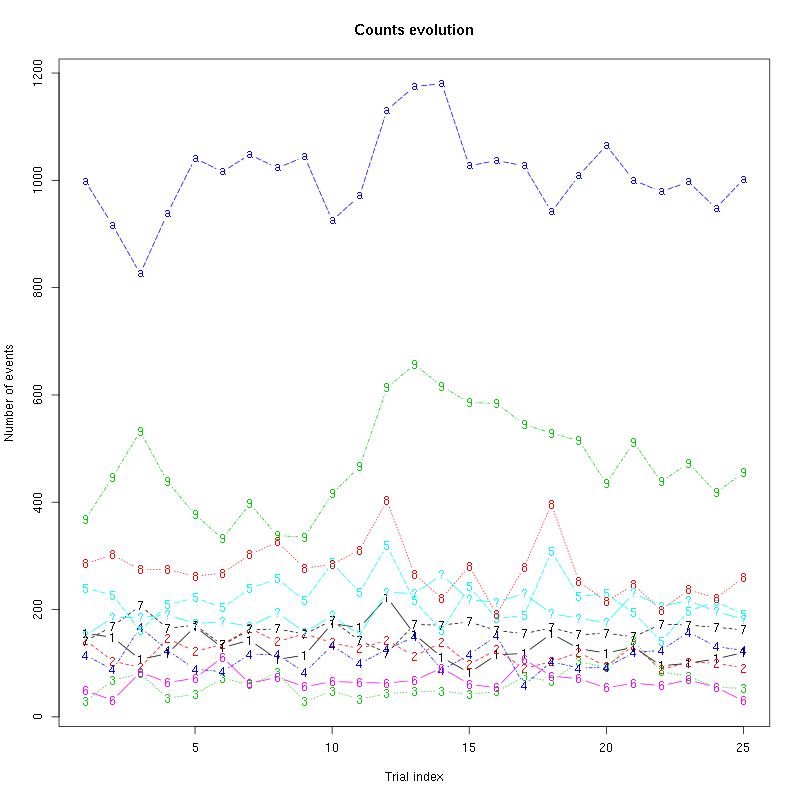
Figure 29: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 25 trials of Vanilla_1 for tetrodeB.
The waveform evolution is:
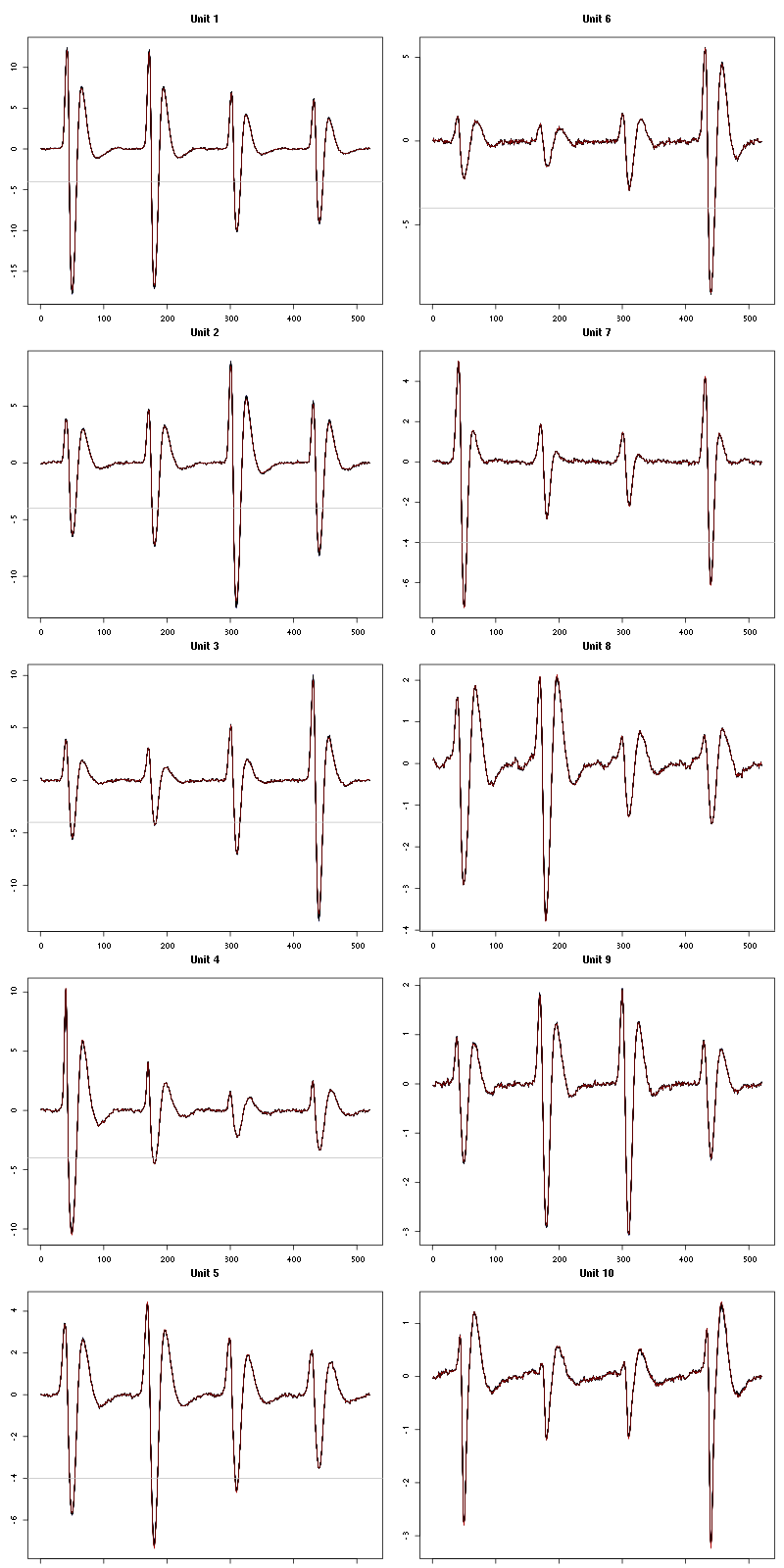
Figure 30: Evolution of the templates of each unit during the 25 trials of Vanilla_1 for tetrodeB.
The observed counting processes, inter spike intervals densities and raster plots are:
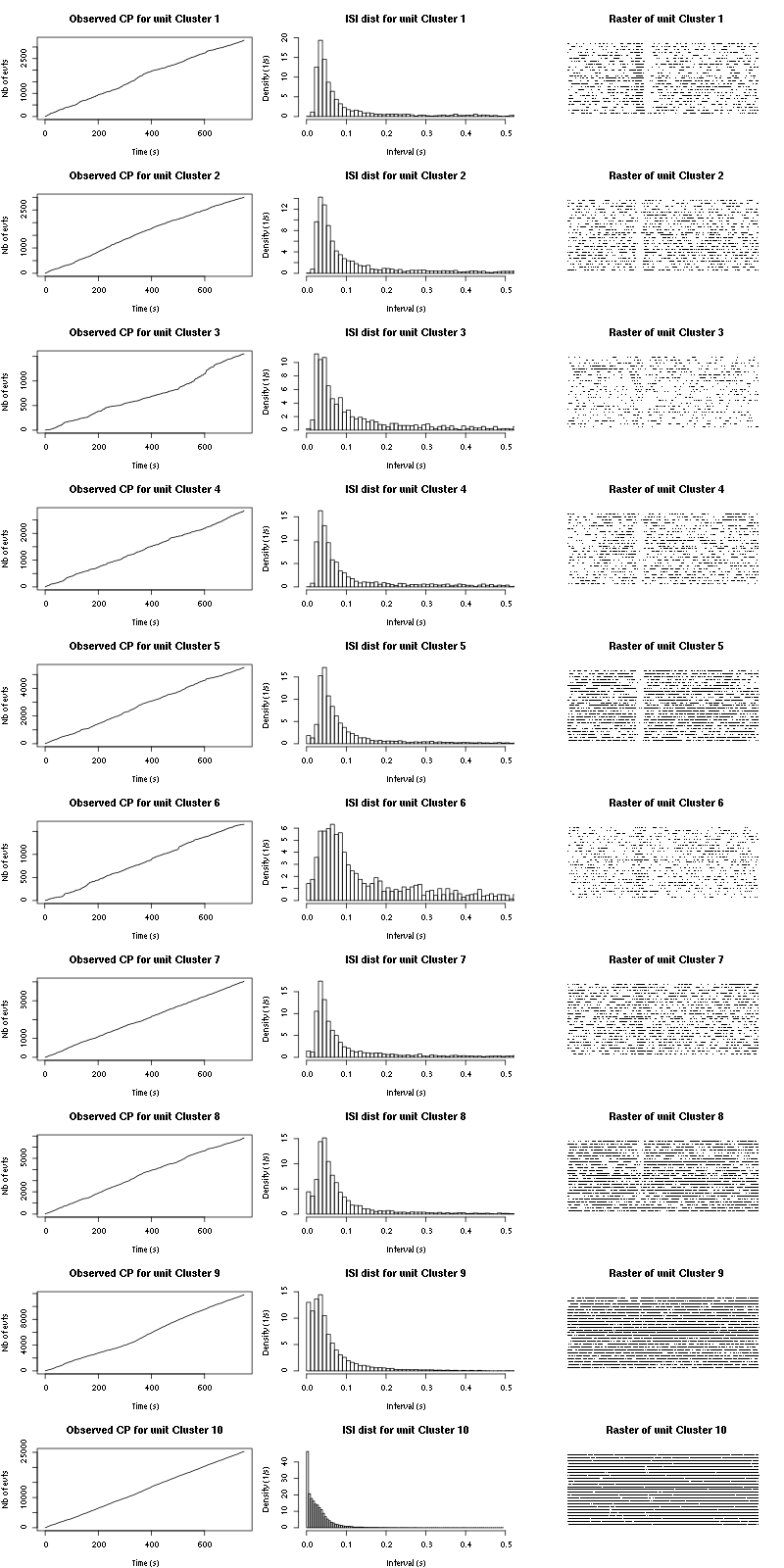
Figure 31: Observed counting processes, empirical inter spike interval distributions and raster plots for Vanilla.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Vanilla_1_tetB$spike_trains[[i]],
a_Vanilla_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
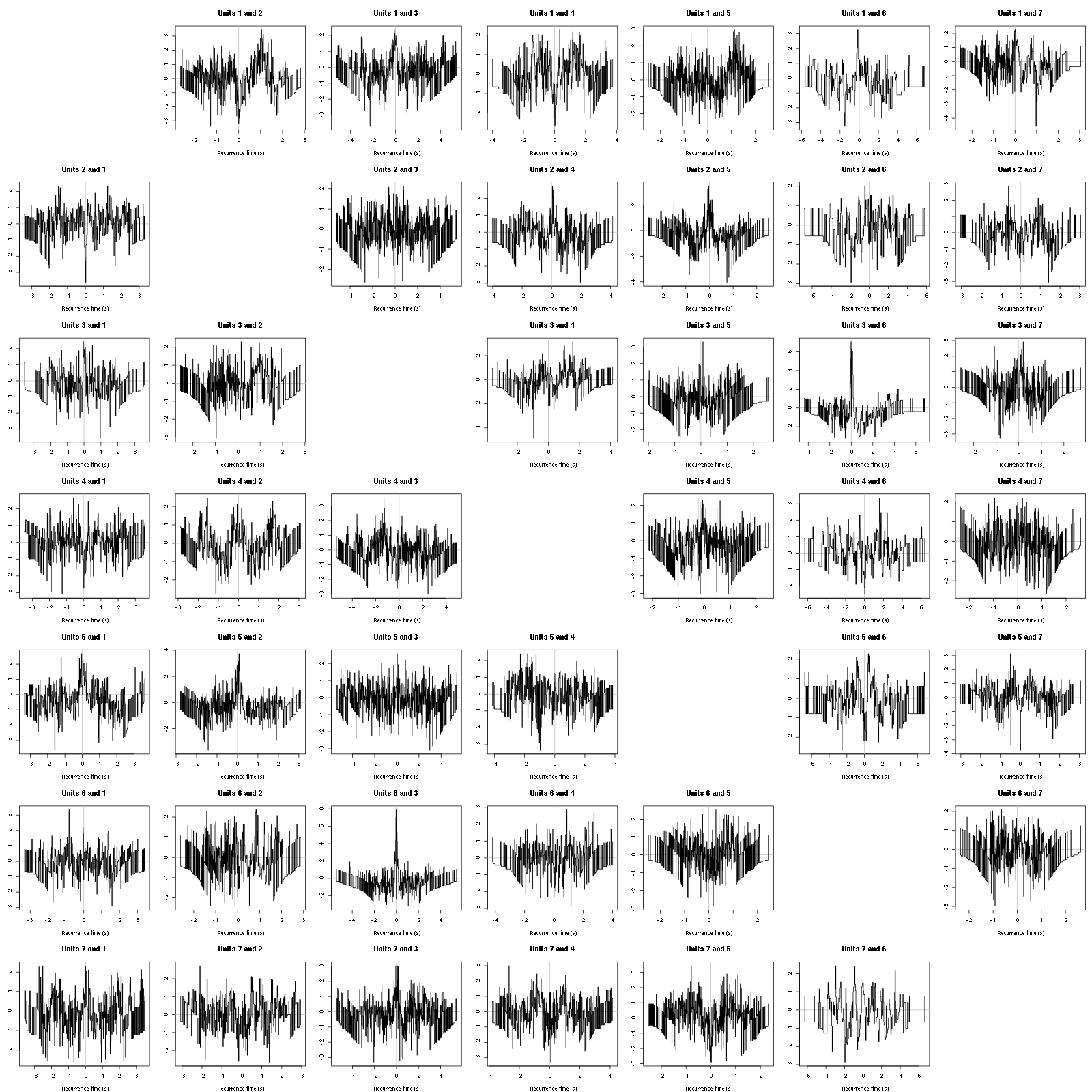
Figure 32: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
8.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Vanilla_1_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Vanilla_1_tetB$spike_trains))
if (!is.null(a_Vanilla_1_tetB$spike_trains[[c_idx]]))
cat(a_Vanilla_1_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Vanilla_1_tetB_u",c_idx,".txt"),sep="\n")
9 25 trials with Octanol
We will carry out an analysis of the 25 trials from Octanol_1. The LabBook mentions the presence of noise in trials 10, 11 and 12, so we skip those.
9.1 Do the job
stim_name = "Octanol_1"
a_Octanol_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=c(1:9,13:25),
centers=a_Vanilla_1_tetB$centers,
counts=a_Vanilla_1_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
9.2 Diagnostic plots
The counts evolution is:
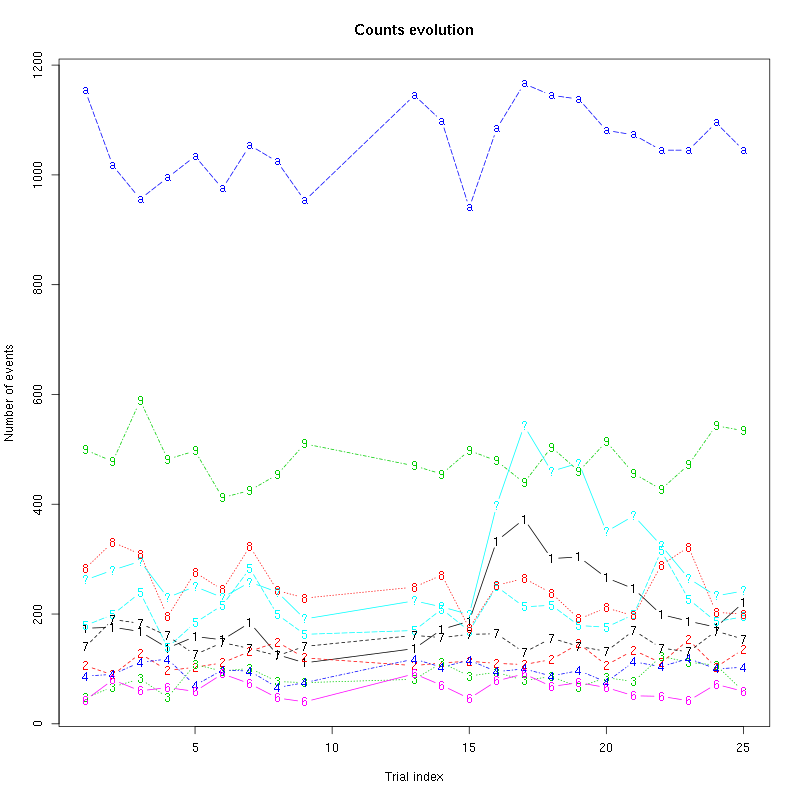
Figure 33: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 25 trials of Octanol_1 for tetrodeB.
The waveform evolution is:
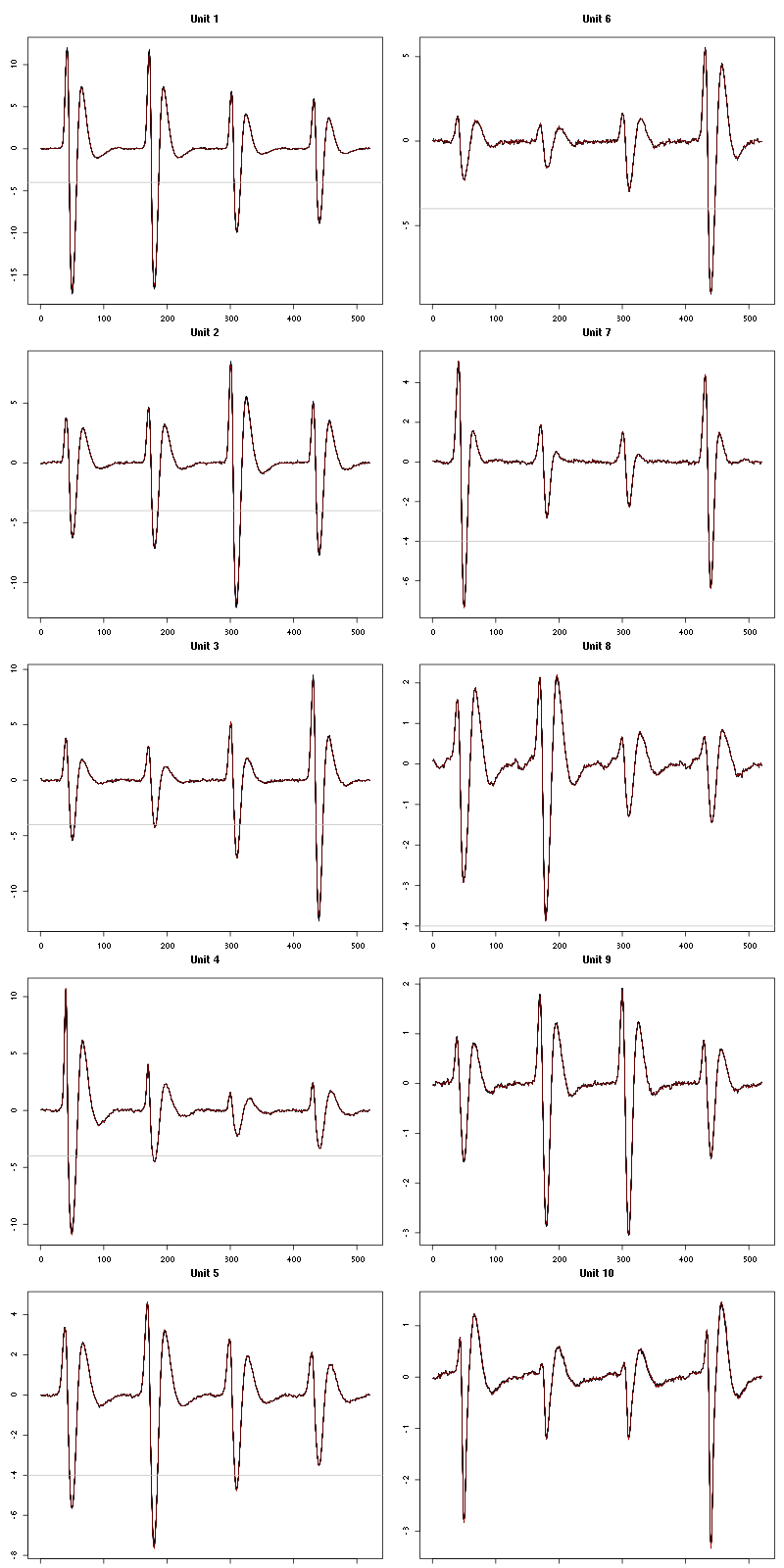
Figure 34: Evolution of the templates of each unit during the 25 trials of Octanol_1 for tetrodeB.
The observed counting processes, inter spike intervals densities and raster plots are:
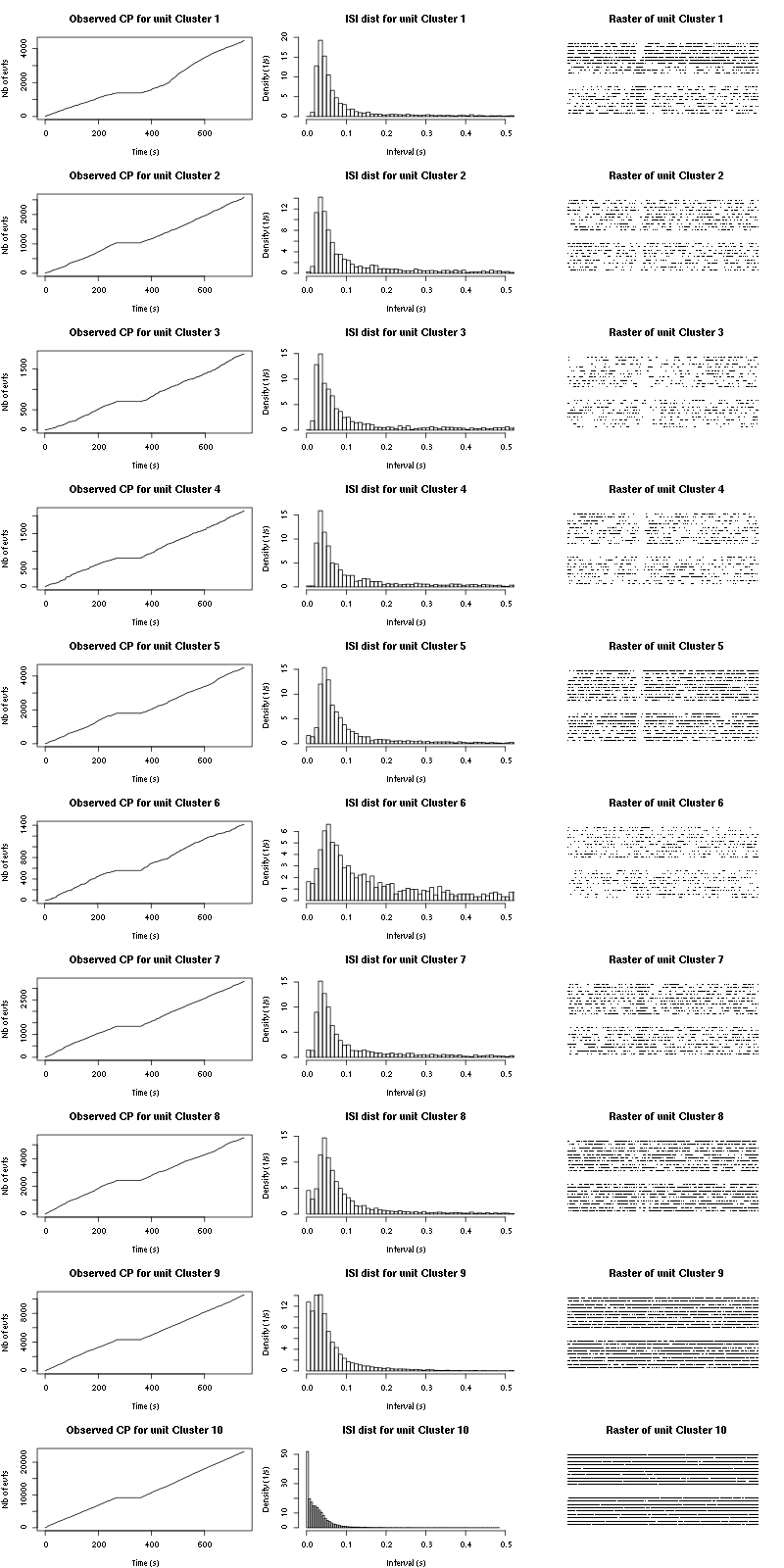
Figure 35: Observed counting processes, empirical inter spike interval distributions and raster plots for Octanol.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Octanol_1_tetB$spike_trains[[i]],
a_Octanol_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
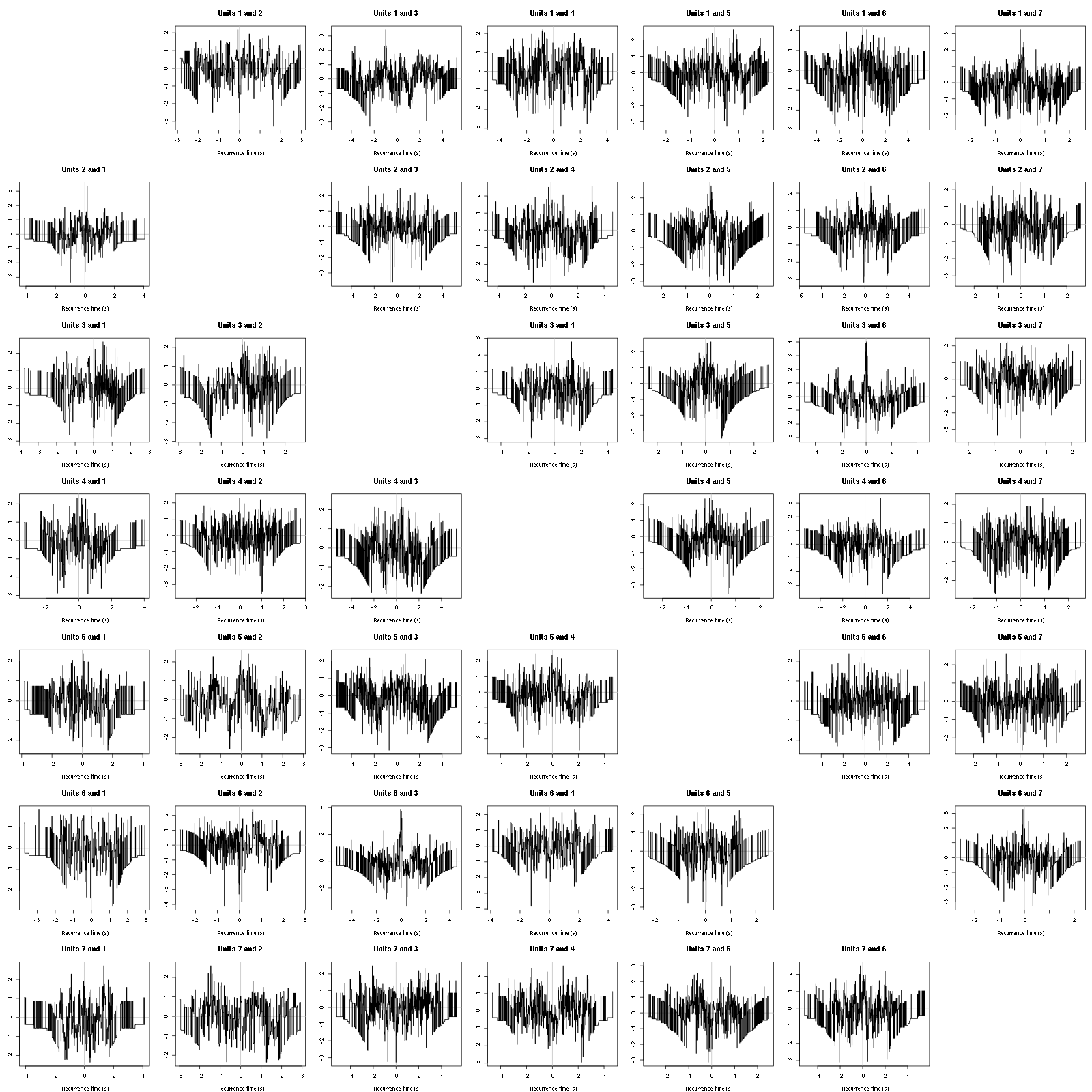
Figure 36: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
9.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Octanol_1_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Octanol_1_tetB$spike_trains))
if (!is.null(a_Octanol_1_tetB$spike_trains[[c_idx]]))
cat(a_Octanol_1_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Octanol_1_tetB_u",c_idx,".txt"),sep="\n")
10 25 trials with Mint
We will carry out an analysis of the 25 trials from Mint_1.
10.1 Do the job
stim_name = "Mint_1"
a_Mint_1_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:25,
centers=a_Octanol_1_tetB$centers,
counts=a_Octanol_1_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
10.2 Diagnostic plots
The counts evolution is:
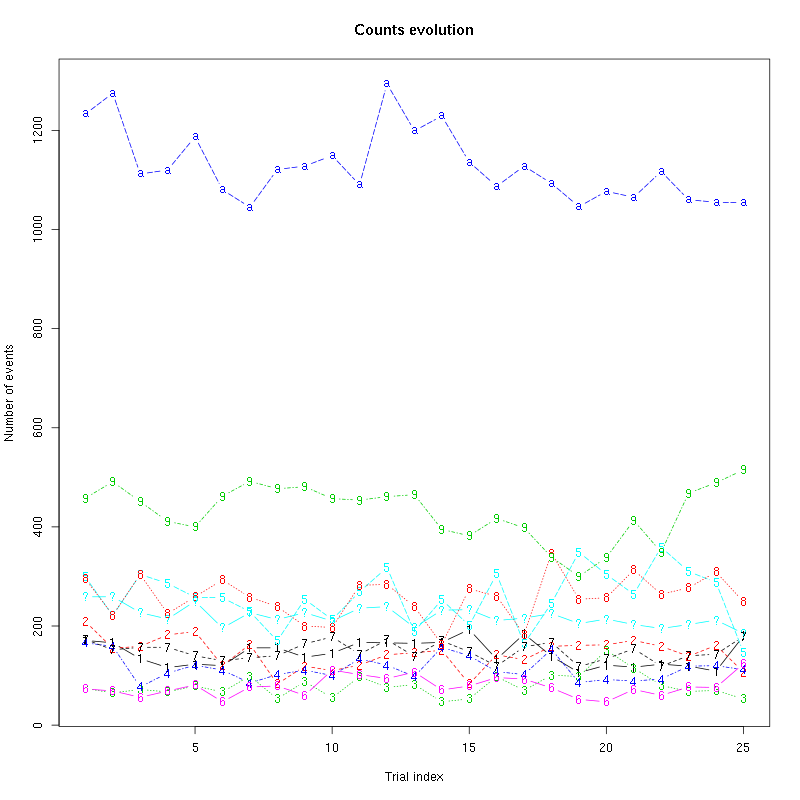
Figure 37: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 25 trials of Mint_1 for tetrodeB.
The waveform evolution is:
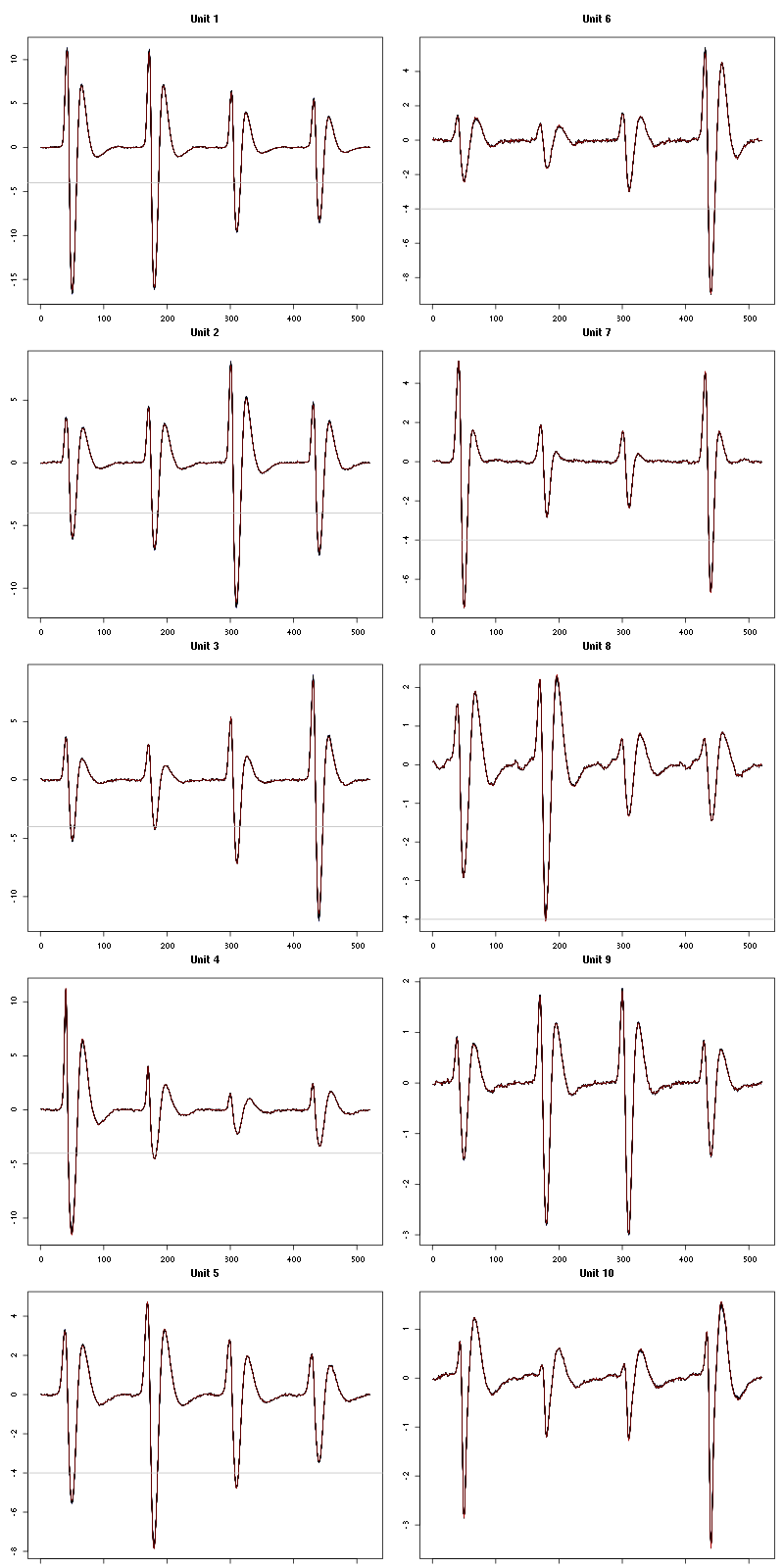
Figure 38: Evolution of the templates of each unit during the 25 trials of Mint_1 for tetrodeB.
The observed counting processes, inter spike intervals densities and raster plots are:
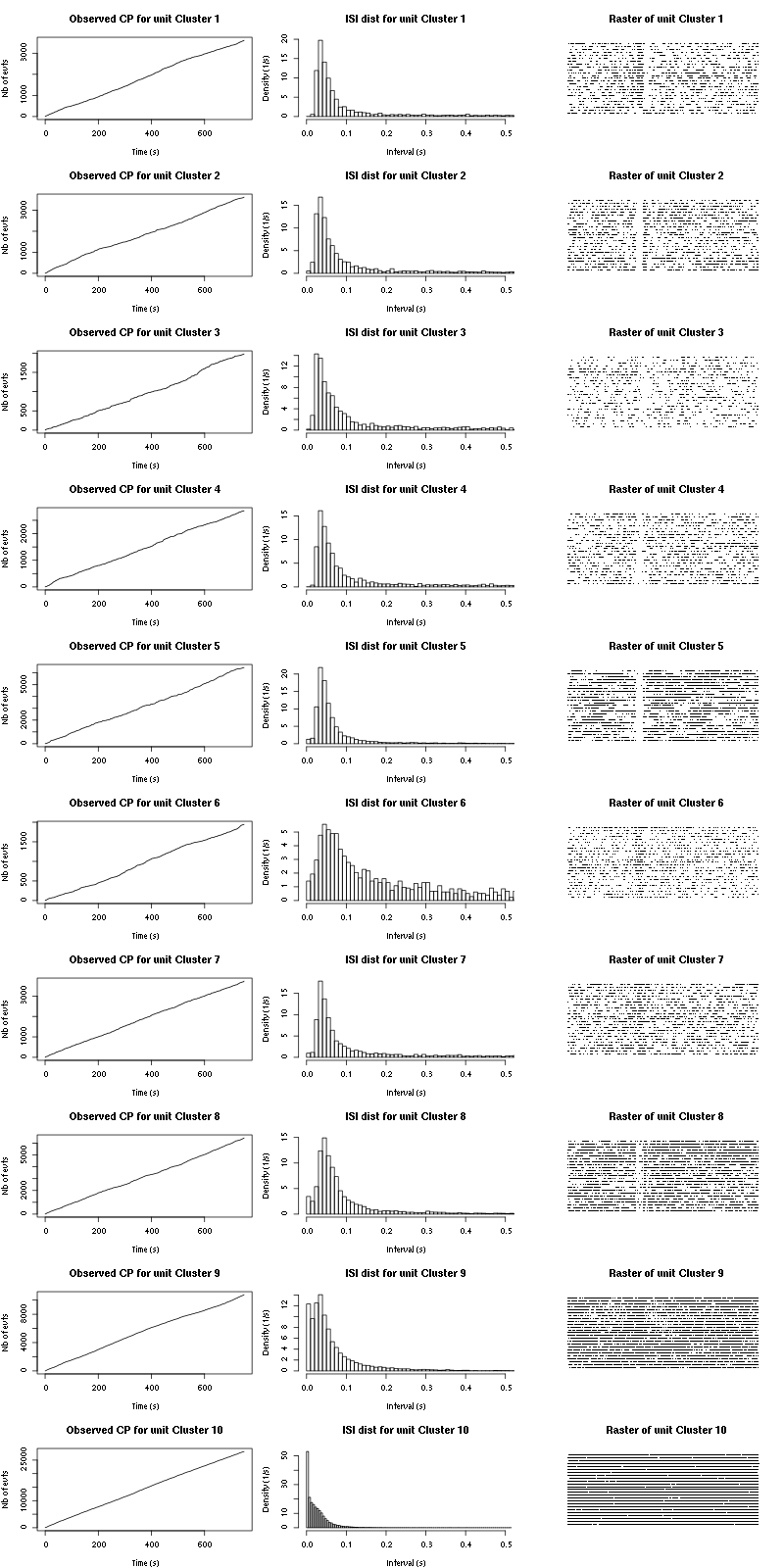
Figure 39: Observed counting processes, empirical inter spike interval distributions and raster plots for Mint.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Mint_1_tetB$spike_trains[[i]],
a_Mint_1_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
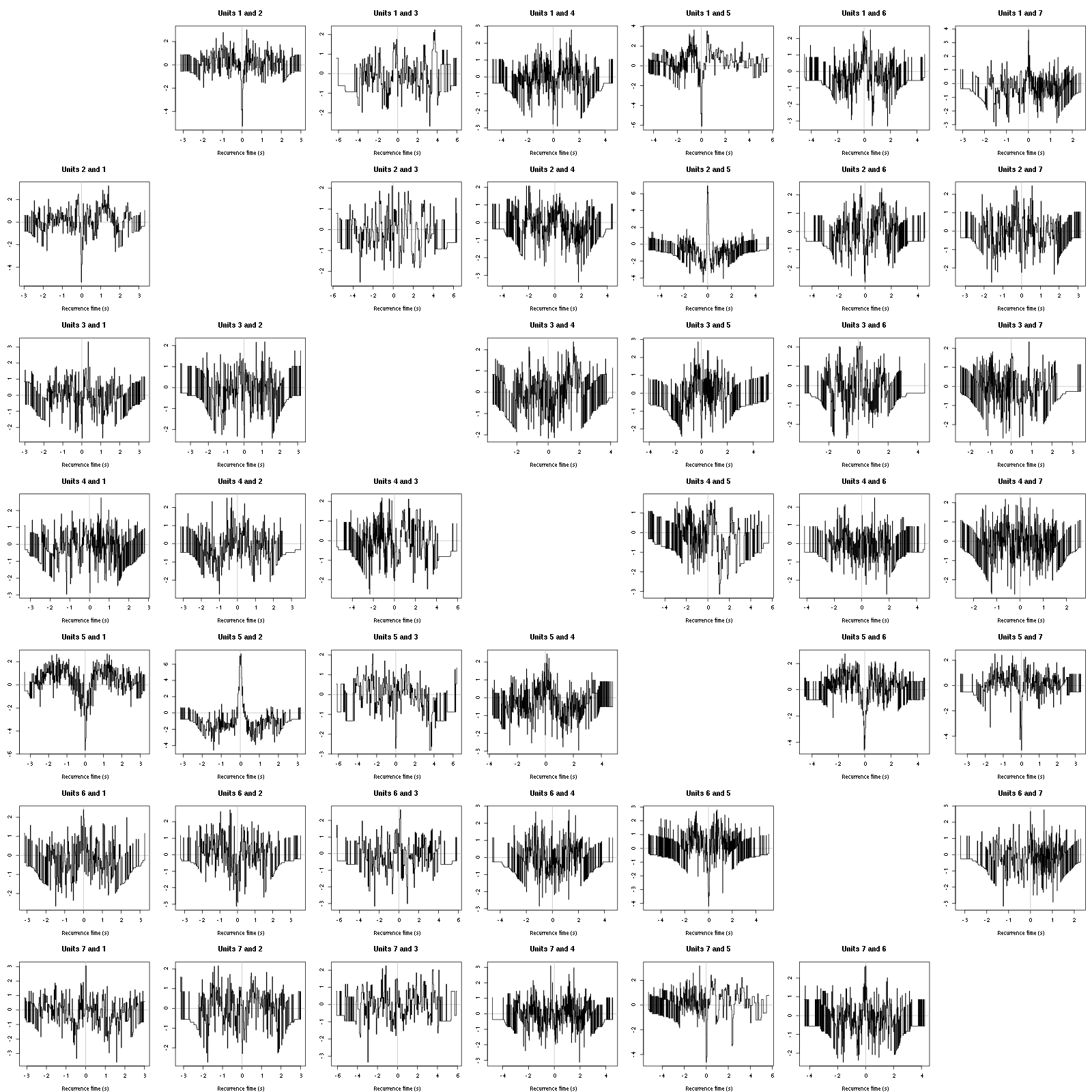
Figure 40: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
10.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Mint_1_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Mint_1_tetB$spike_trains))
if (!is.null(a_Mint_1_tetB$spike_trains[[c_idx]]))
cat(a_Mint_1_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Mint_1_tetB_u",c_idx,".txt"),sep="\n")
11 25 trials with C3H_2
We will carry out an analysis of the second set of 25 trials with Cis-3-Hexen-1-ol from C3H_2.
11.1 Do the job
stim_name = "C3H_2"
a_C3H_2_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:25,
centers=a_Mint_1_tetB$centers,
counts=a_Mint_1_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
11.2 Diagnostic plots
The counts evolution is:
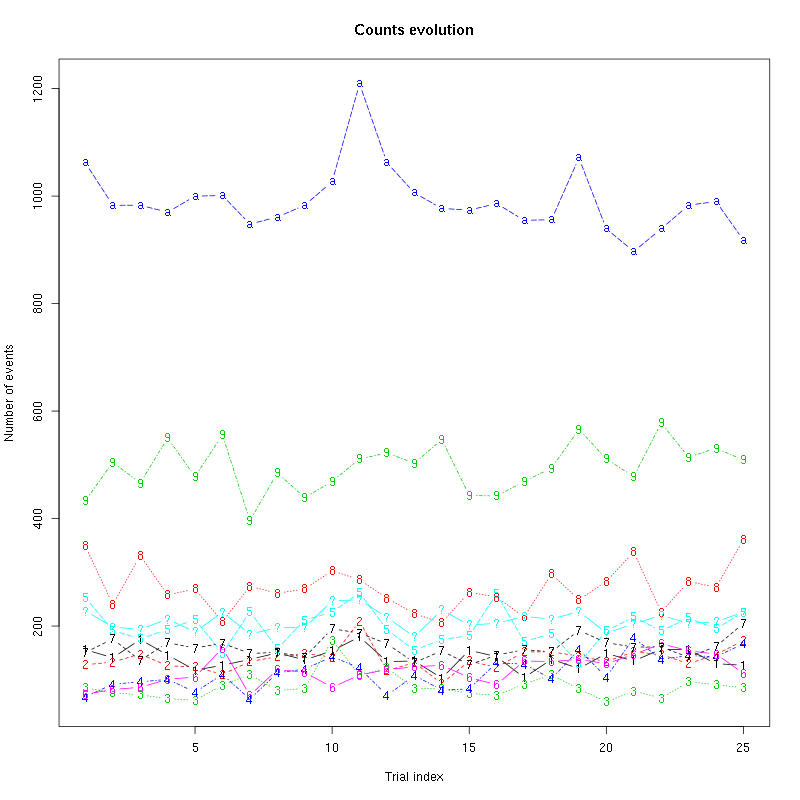
Figure 41: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 25 trials of C3H_2 for tetrodeB.
The waveform evolution is:
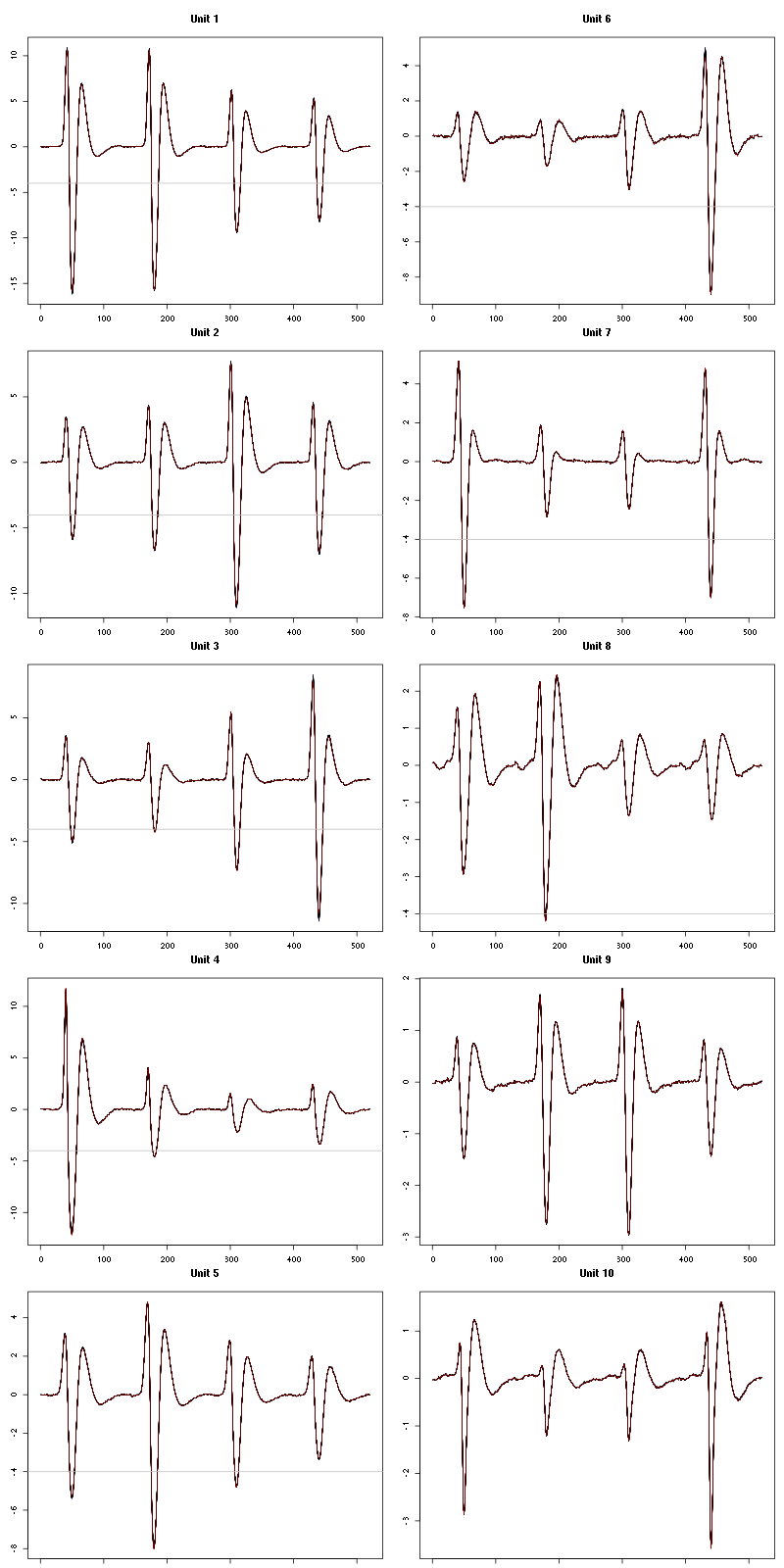
Figure 42: Evolution of the templates of each unit during the 25 trials of C3H_2 for tetrodeB.
The observed counting processes, inter spike intervals densities and raster plots are:
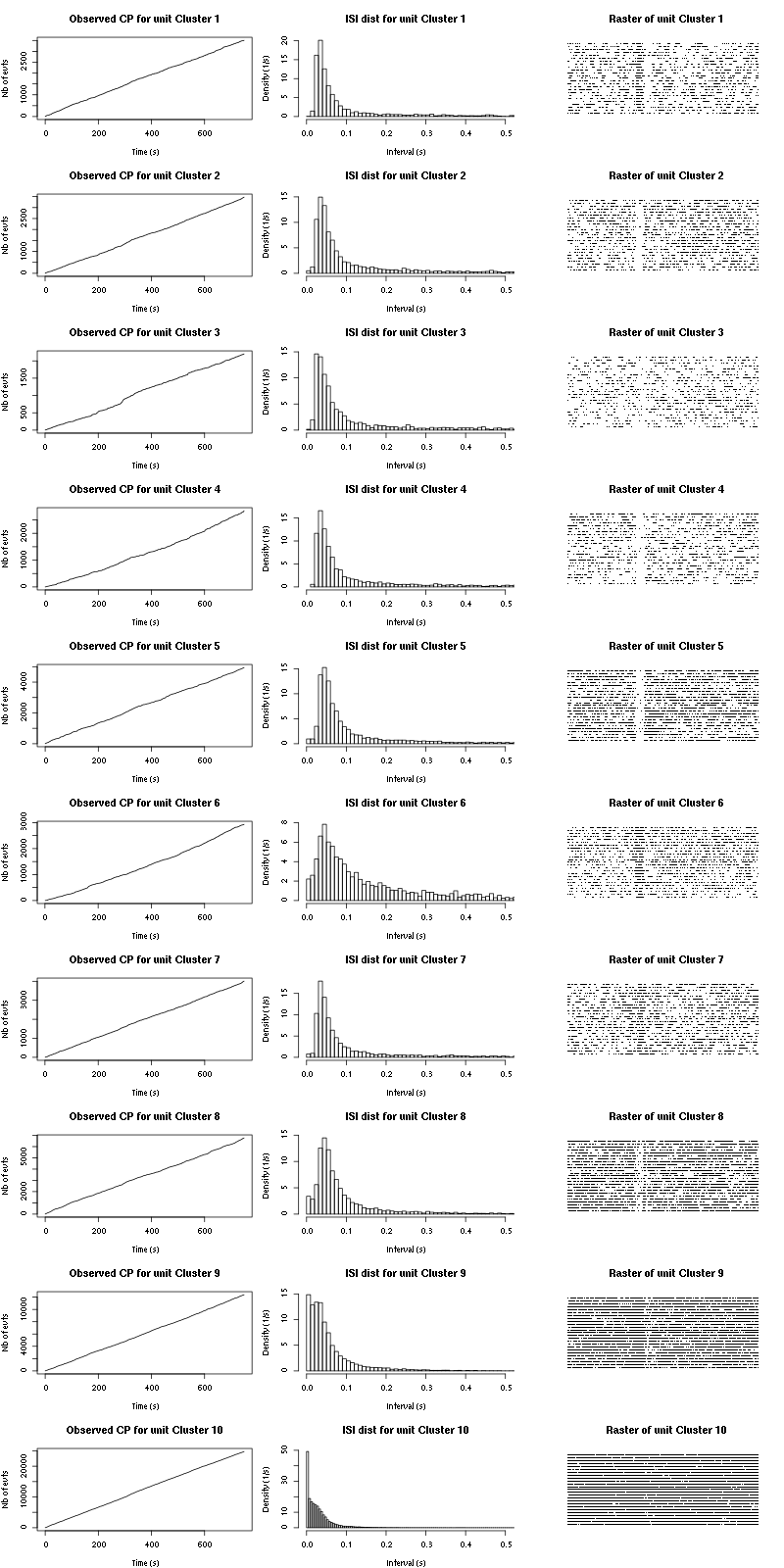
Figure 43: Observed counting processes, empirical inter spike interval distributions and raster plots for Mint.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_2_tetB$spike_trains[[i]],
a_C3H_2_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
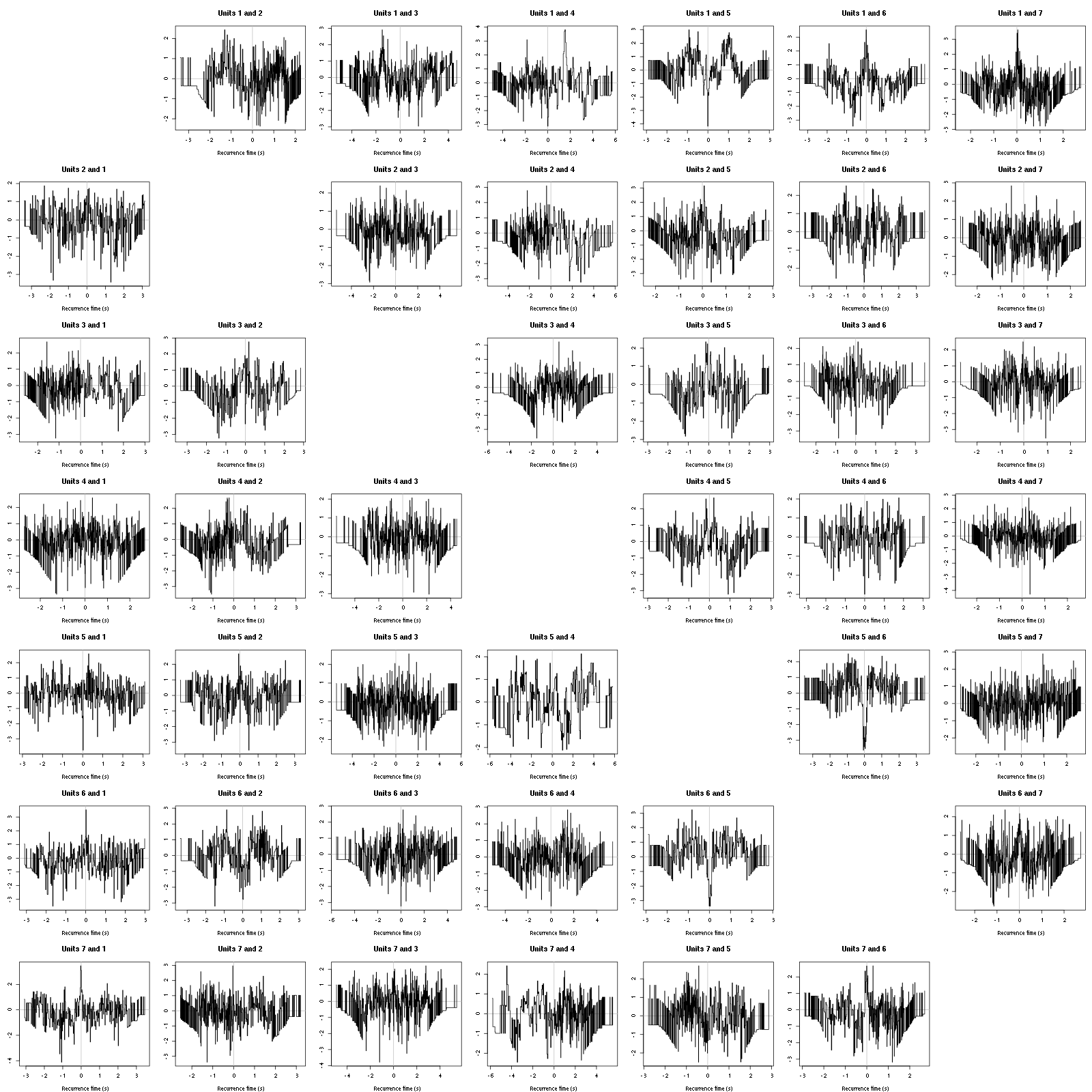
Figure 44: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
11.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_2_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_2_tetB$spike_trains))
if (!is.null(a_C3H_2_tetB$spike_trains[[c_idx]]))
cat(a_C3H_2_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_C3H_2_tetB_u",c_idx,".txt"),sep="\n")
12 30 trials of Spontaneous_3
We must redefine our get_tetrode_data function since the HDF5 containing the data is not the one where the previous data were stored:
12.1 Redefine get_tetrode_data
get_tetrode_data = function(trial_idx,stim="Spontaneous_1") {
prefix = ifelse(trial_idx<10,
paste0("/",stim,"/trial_0",trial_idx),
paste0("/",stim,"/trial_",trial_idx)
)
cbind(h5read("locust20010214_part2.hdf5", paste0(prefix,"/ch02")),
h5read("locust20010214_part2.hdf5", paste0(prefix,"/ch03")),
h5read("locust20010214_part2.hdf5", paste0(prefix,"/ch05")),
h5read("locust20010214_part2.hdf5", paste0(prefix,"/ch07")))
}
12.2 Do the job
stim_name = "Spontaneous_3"
a_Spontaneous_3_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:30,
centers=a_C3H_2_tetB$centers,
counts=a_C3H_2_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
12.3 Diagnostic plots
The counts evolution is:
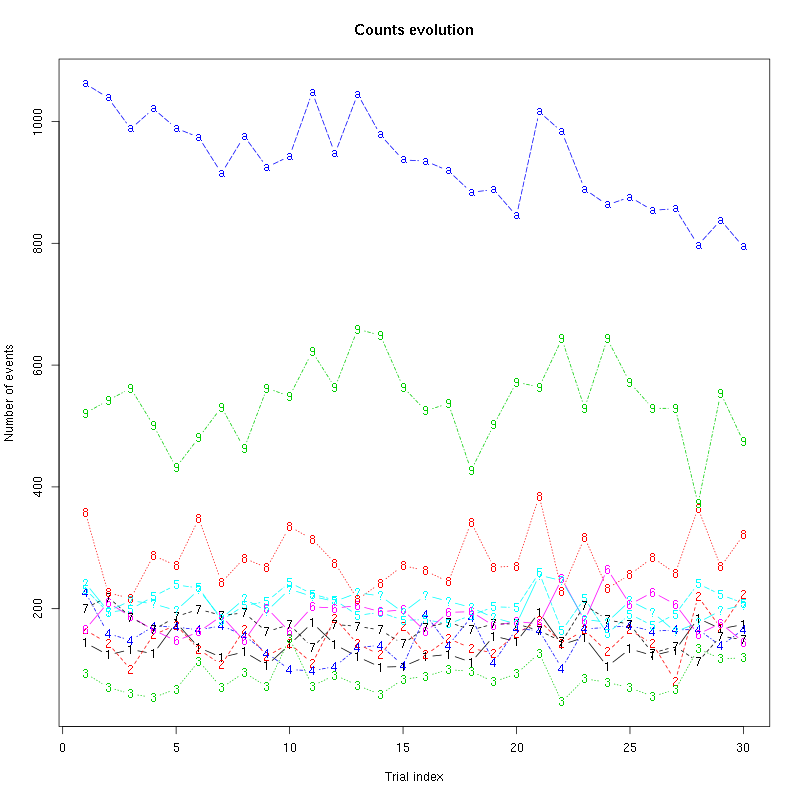
Figure 45: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of Spontaneous_3 for tetrodeB.
The waveform evolution is:
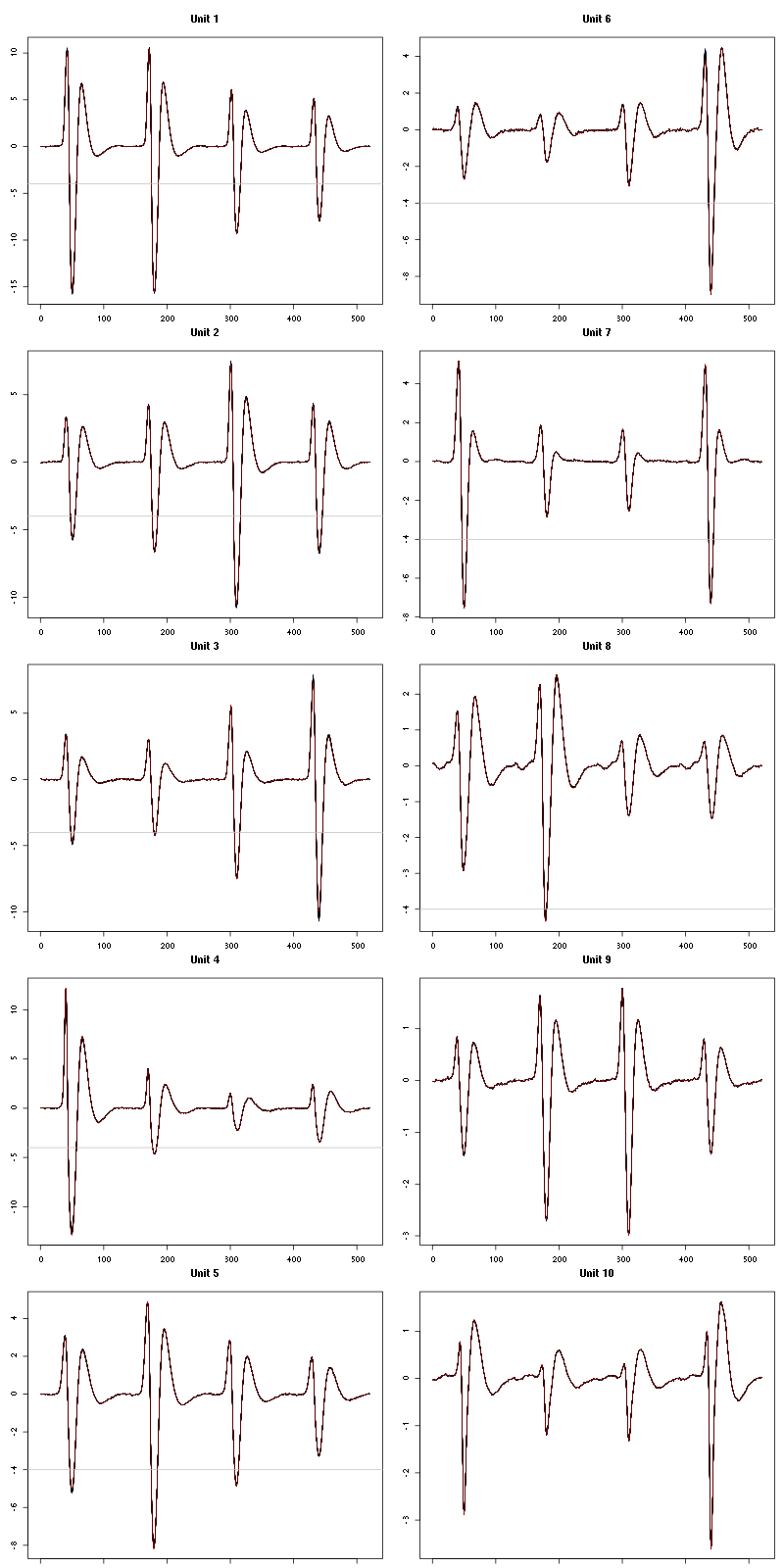
Figure 46: Evolution of the templates of each unit during the 30 trials of Spontaneous_3 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
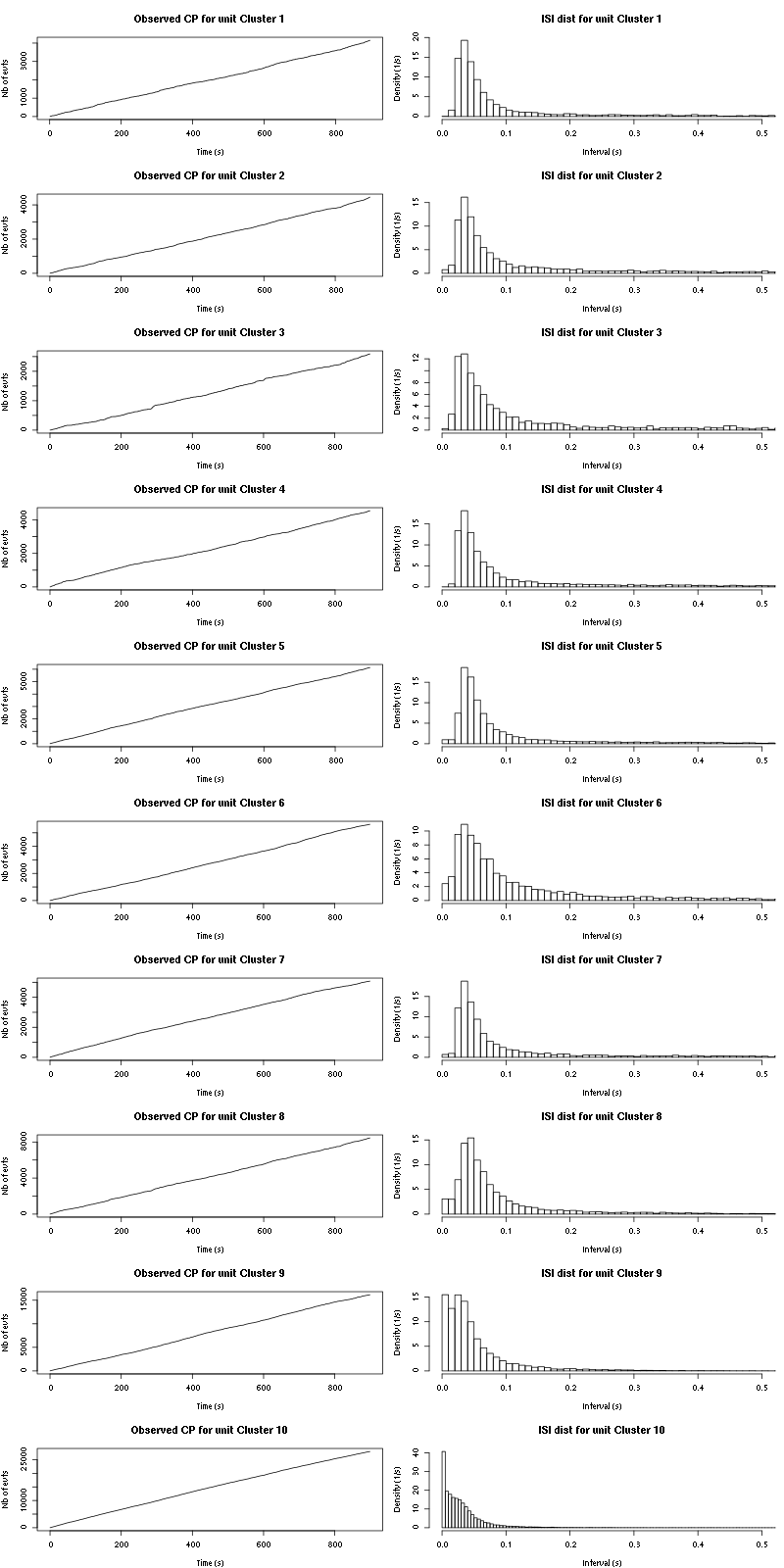
Figure 47: Observed counting processes, empirical inter spike interval distributions and raster plots for Spontaneous_3.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Spontaneous_3_tetB$spike_trains[[i]],
a_Spontaneous_3_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
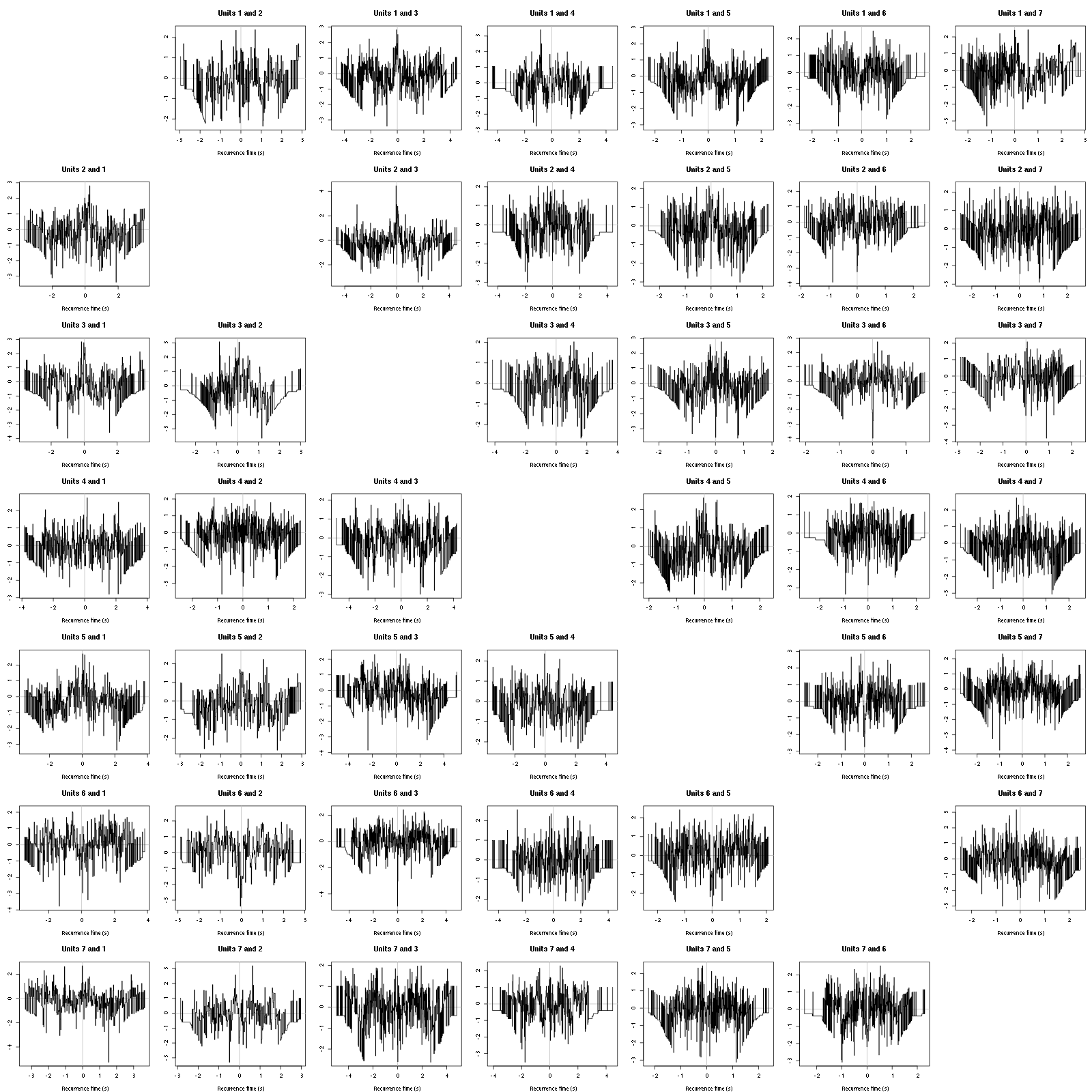
Figure 48: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
12.4 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Spontaneous_3_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Spontaneous_3_tetB$spike_trains))
if (!is.null(a_Spontaneous_3_tetB$spike_trains[[c_idx]]))
cat(a_Spontaneous_3_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Spontaneous_3_tetB_u",c_idx,".txt"),sep="\n")
13 30 trials of Spontaneous_4
We now analyze the 30 trials from Spontaneous_4.
13.1 Do the job
stim_name = "Spontaneous_4"
a_Spontaneous_4_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:30,
centers=a_Spontaneous_3_tetB$centers,
counts=a_Spontaneous_3_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
13.2 Diagnostic plots
The counts evolution is:
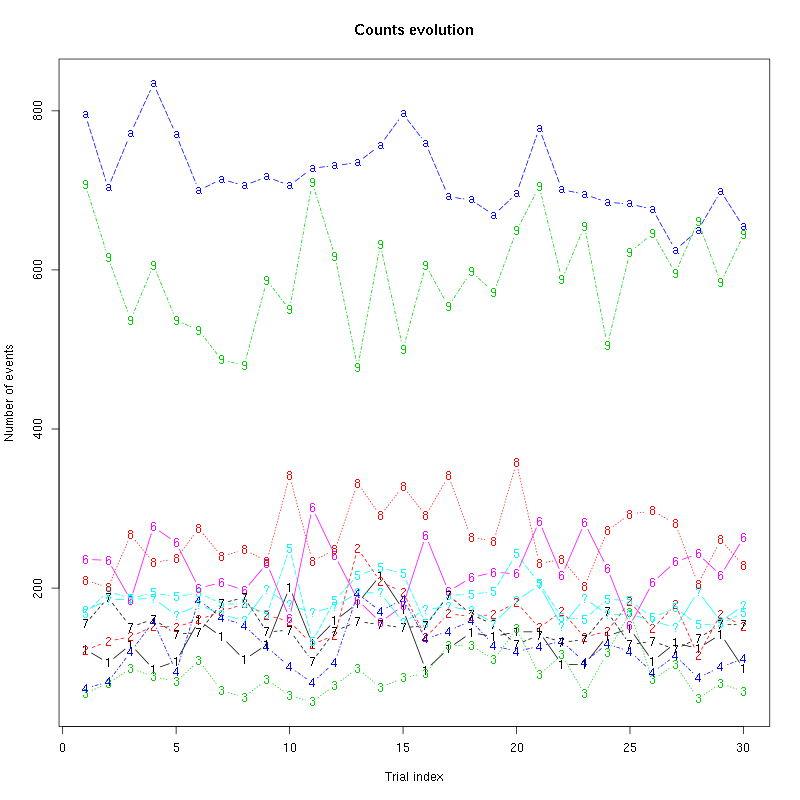
Figure 49: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of Spontaneous_4 for tetrodeB.
The waveform evolution is:
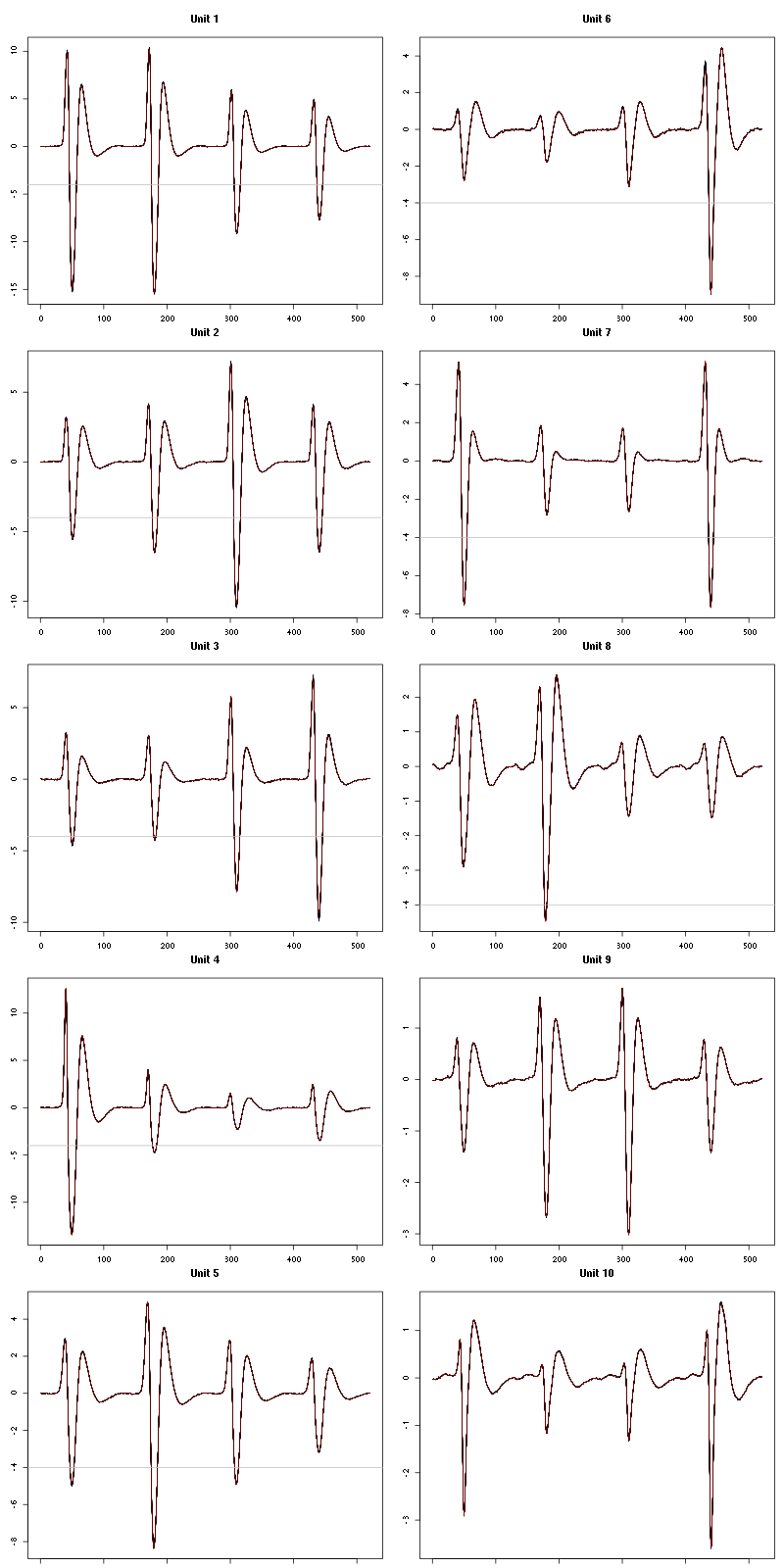
Figure 50: Evolution of the templates of each unit during the 30 trials of Spontaneous_4 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
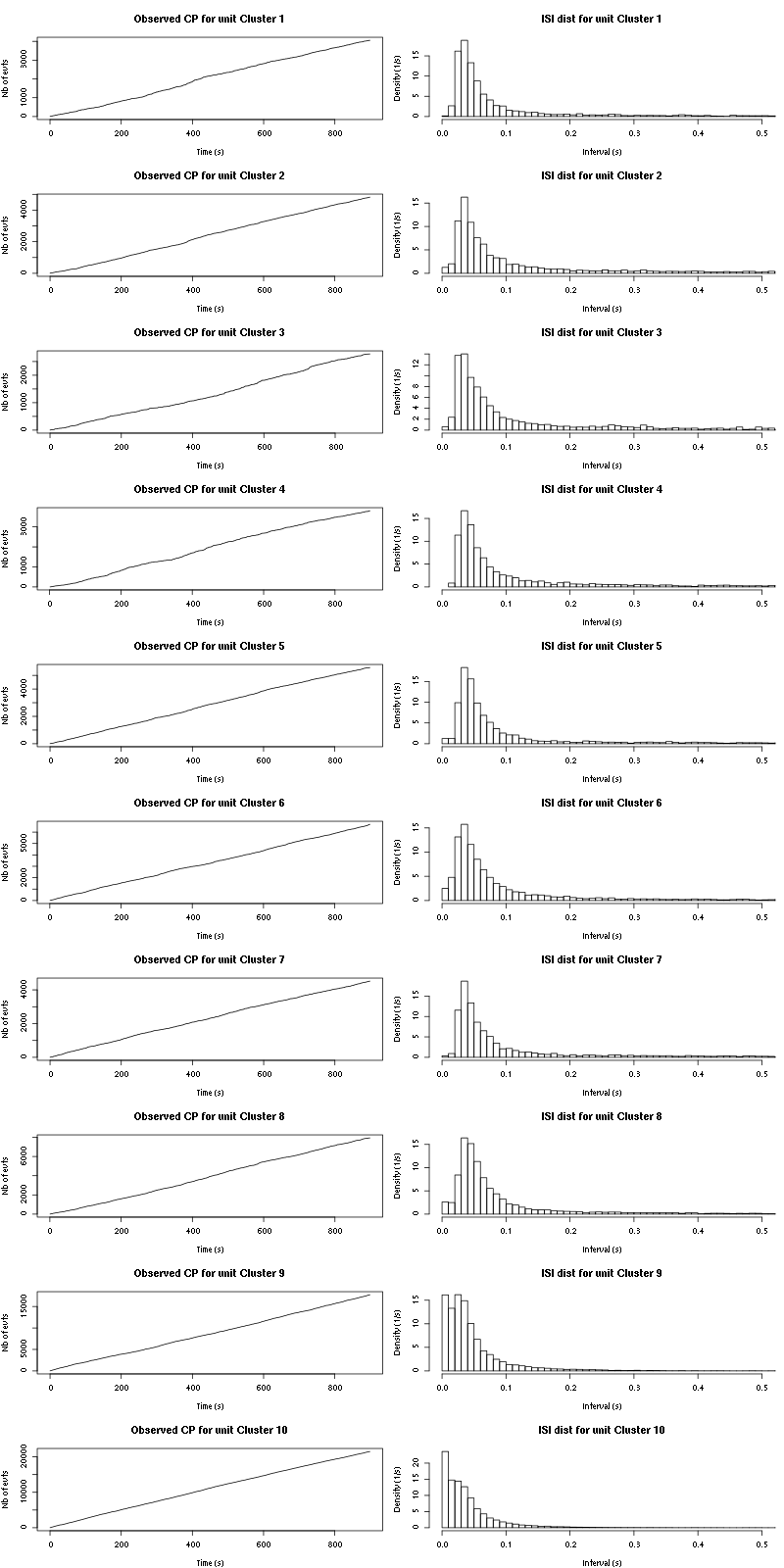
Figure 51: Observed counting processes, empirical inter spike interval distributions and raster plots for Spontaneous_4.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_Spontaneous_4_tetB$spike_trains[[i]],
a_Spontaneous_4_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
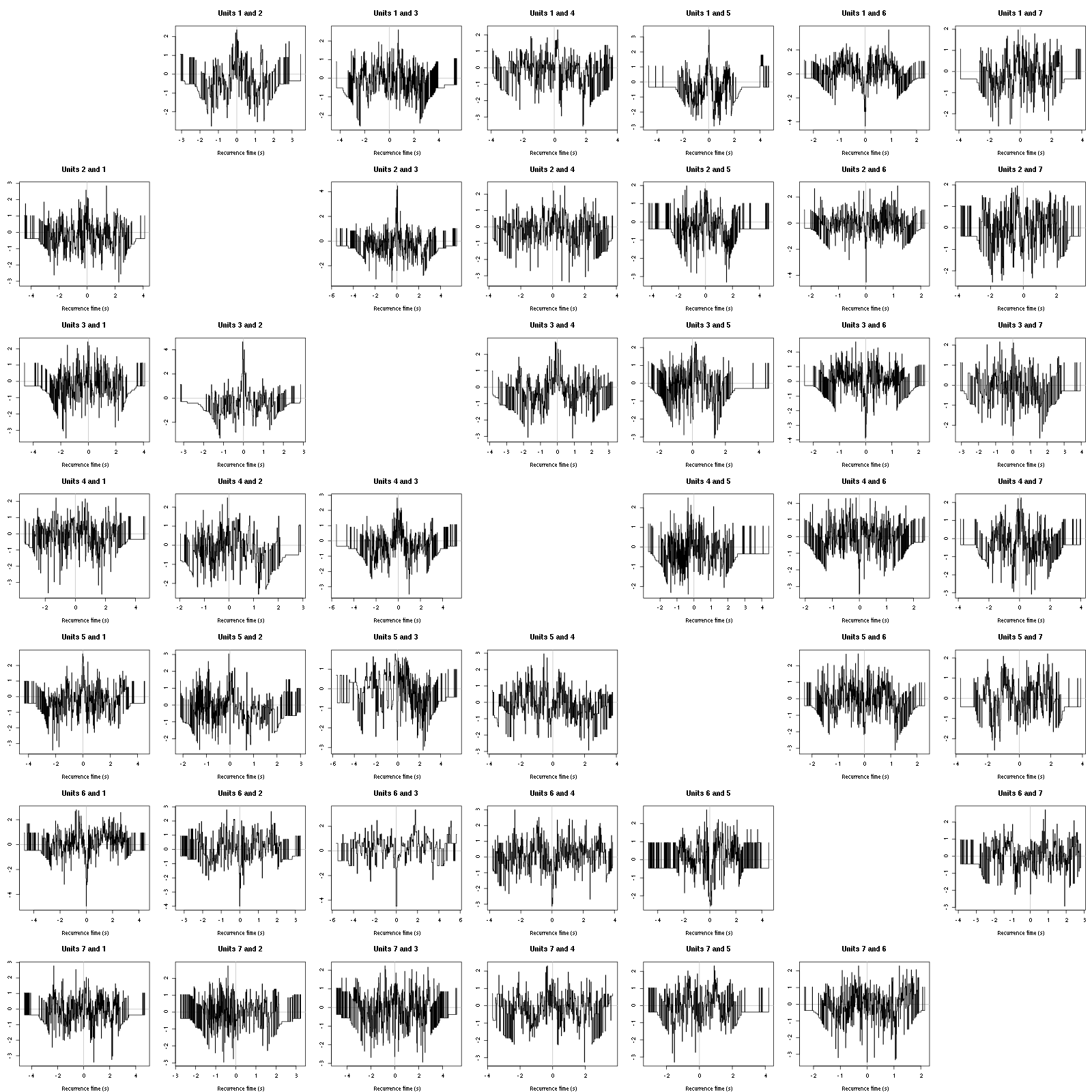
Figure 52: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
13.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_Spontaneous_4_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_Spontaneous_4_tetB$spike_trains))
if (!is.null(a_Spontaneous_4_tetB$spike_trains[[c_idx]]))
cat(a_Spontaneous_4_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_Spontaneous_4_tetB_u",c_idx,".txt"),sep="\n")
14 30 trials of C3H_3
14.1 Do the job
stim_name = "C3H_3"
a_C3H_3_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:30,
centers=a_Spontaneous_4_tetB$centers,
counts=a_Spontaneous_4_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
14.2 Diagnostic plots
The counts evolution is:
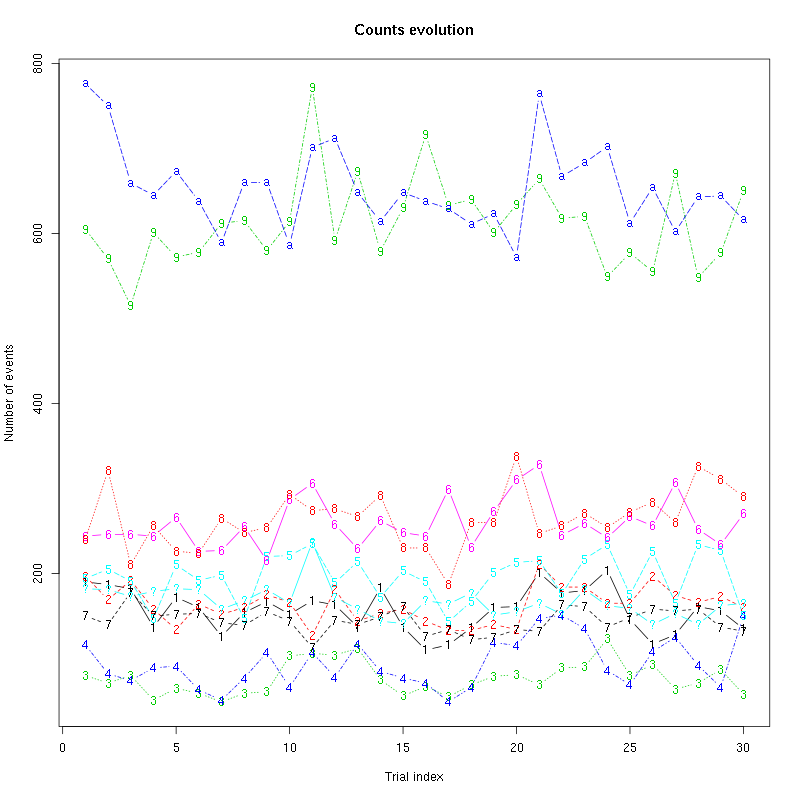
Figure 53: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of C3H_3 for tetrodeB.
The waveform evolution is:
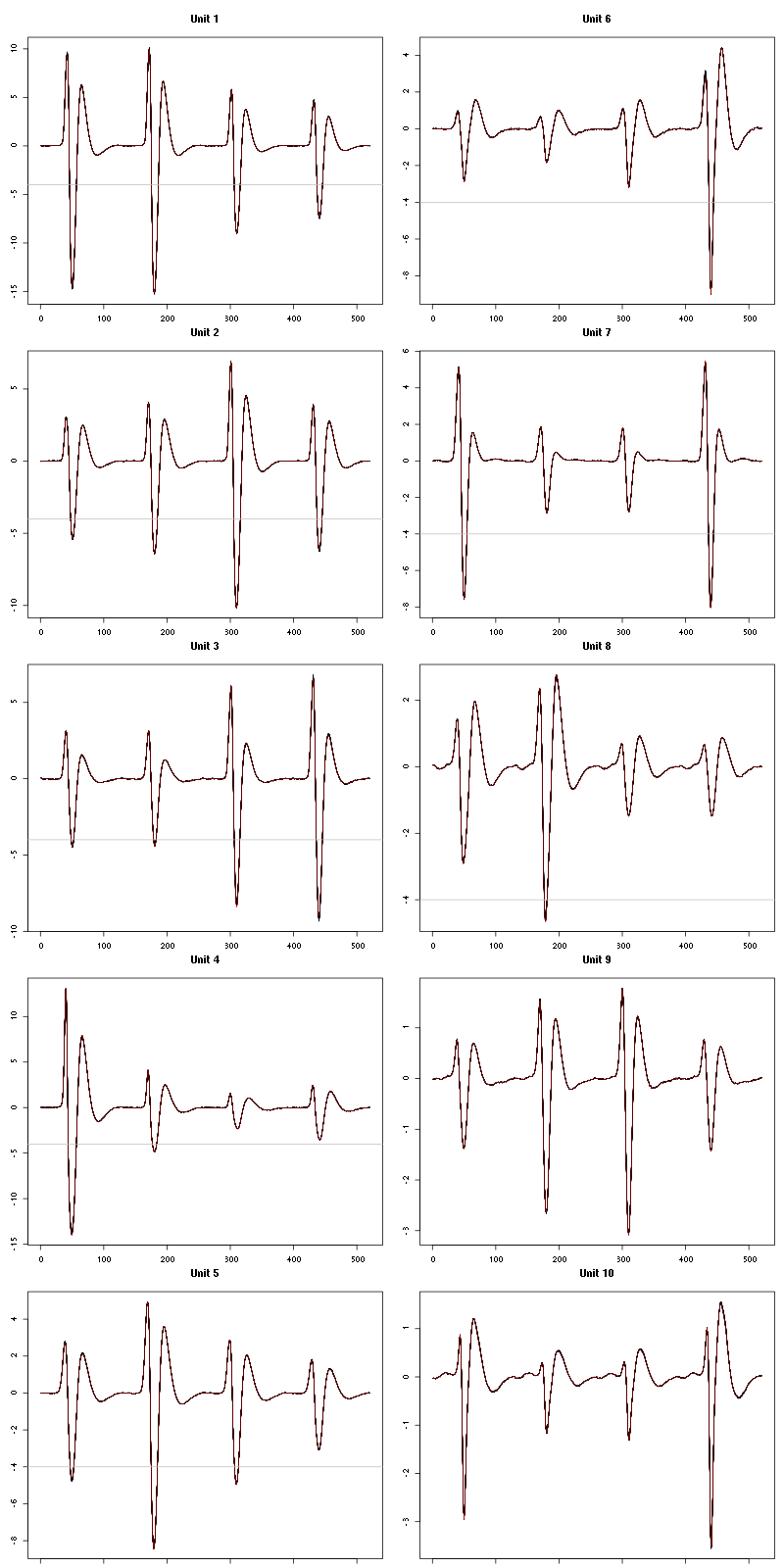
Figure 54: Evolution of the templates of each unit during the 30 trials of C3H_3 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
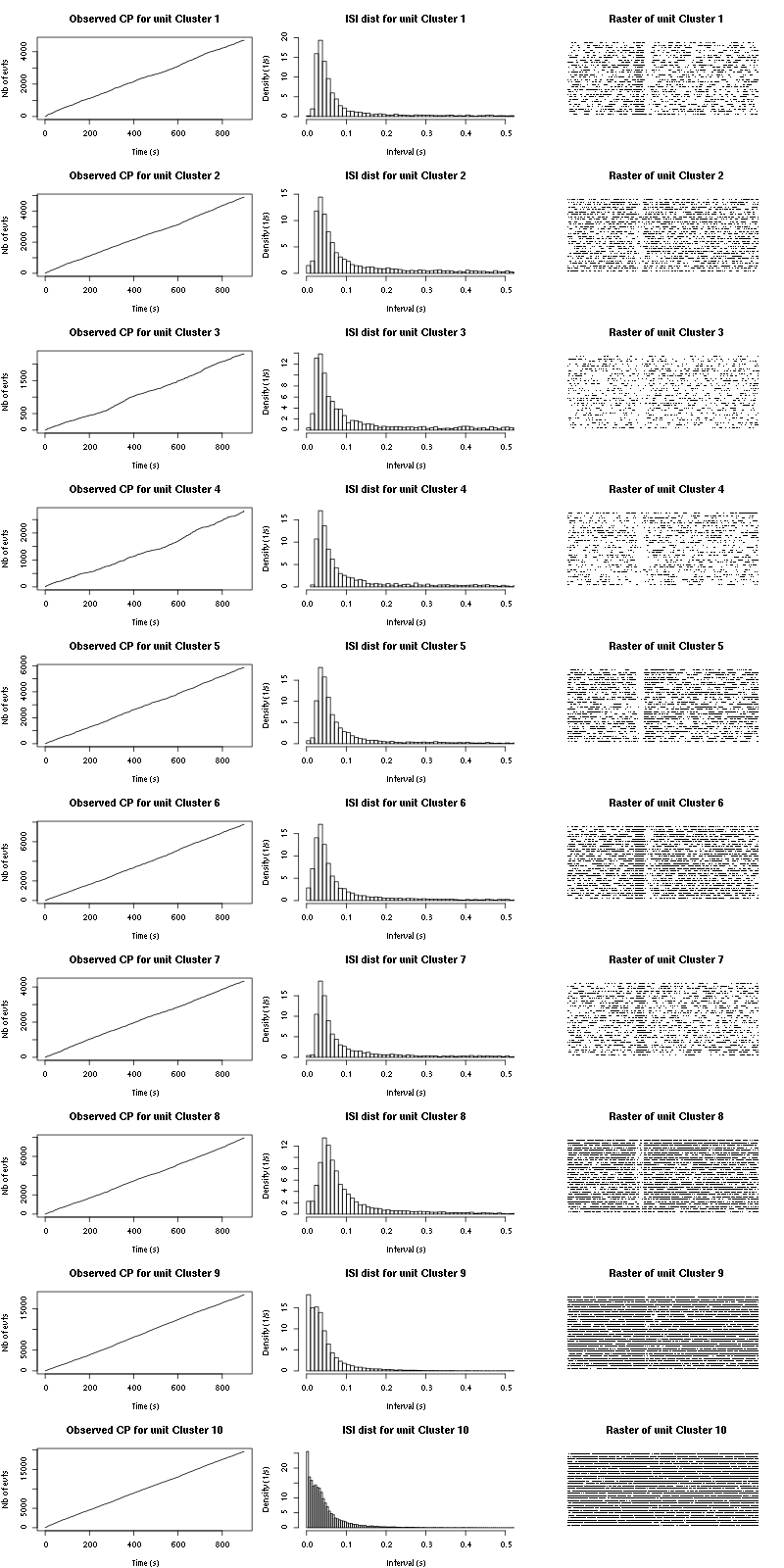
Figure 55: Observed counting processes, empirical inter spike interval distributions and raster plots for C3H_3.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_3_tetB$spike_trains[[i]],
a_C3H_3_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
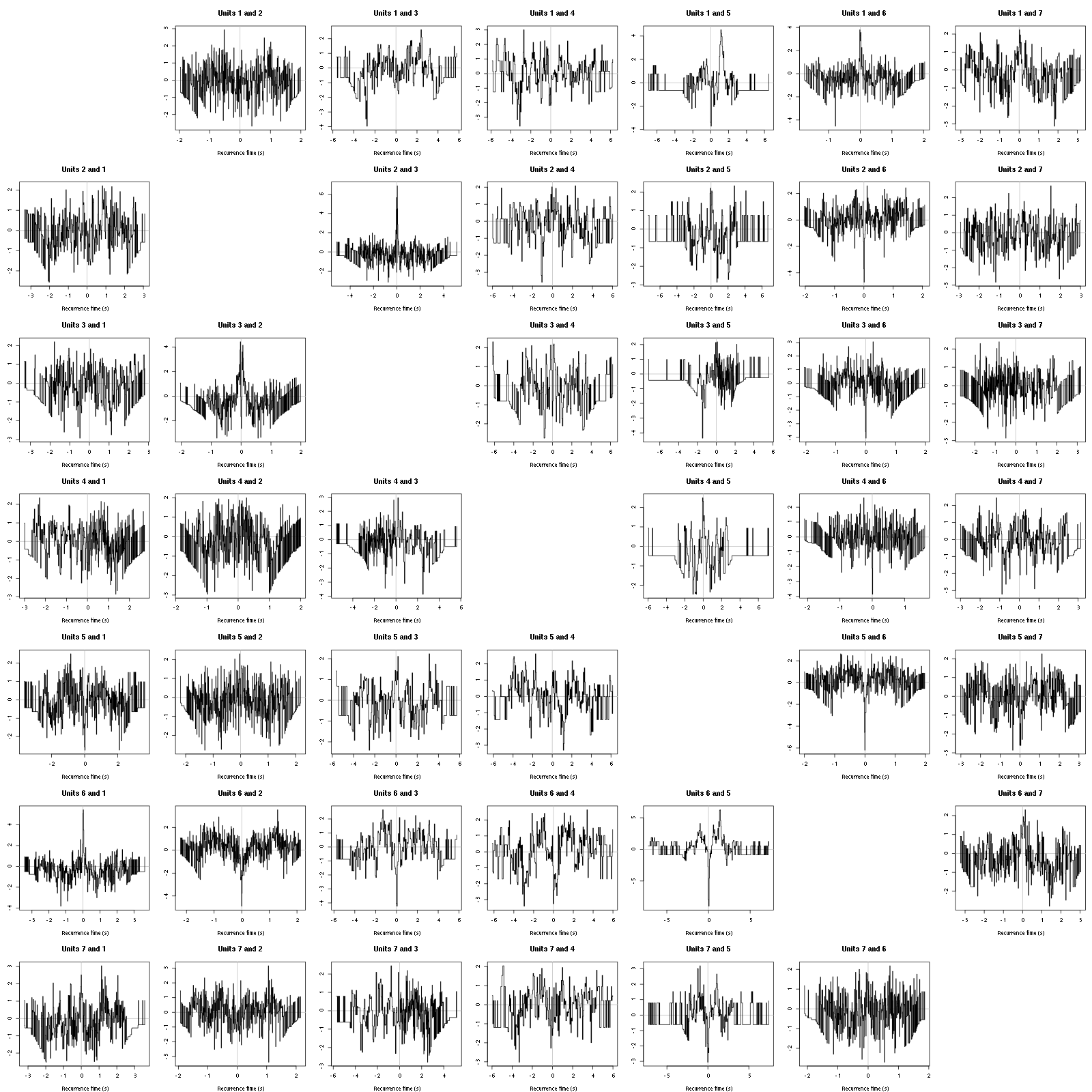
Figure 56: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
14.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_3_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_3_tetB$spike_trains))
if (!is.null(a_C3H_3_tetB$spike_trains[[c_idx]]))
cat(a_C3H_3_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_C3H_3_tetB_u",c_idx,".txt"),sep="\n")
15 10 trials of C3H_4
The LabBook mentions that the disk got filled up during trial 11 (30 were planed ) so we stop at trial 10.
15.1 Do the job
stim_name = "C3H_4"
a_C3H_4_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:10,
centers=a_C3H_3_tetB$centers,
counts=a_C3H_3_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
15.2 Diagnostic plots
The counts evolution is:
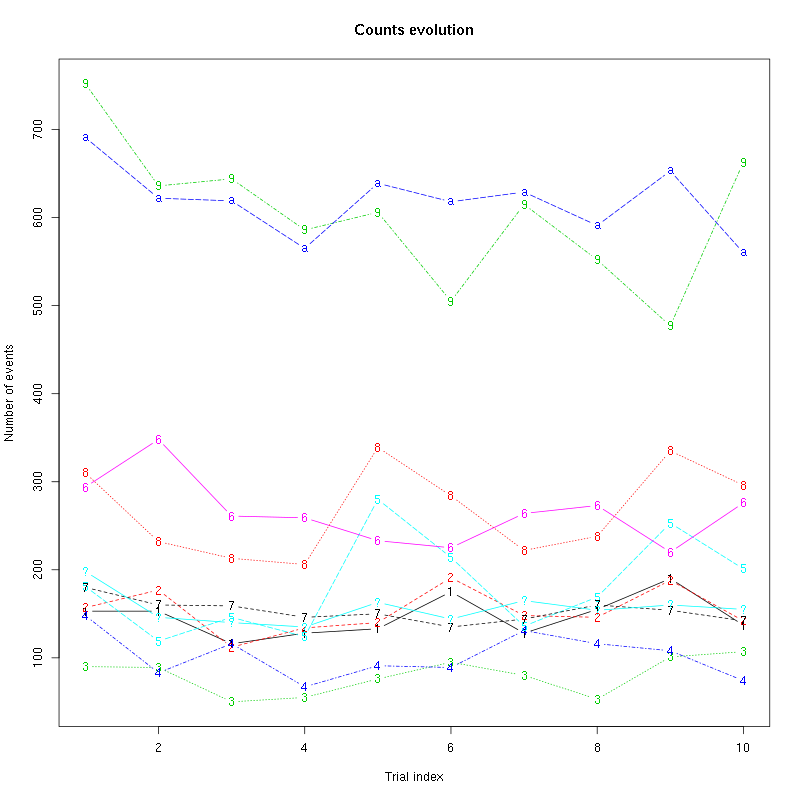
Figure 57: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 10 trials of C3H_4 for tetrodeB.
The waveform evolution is:
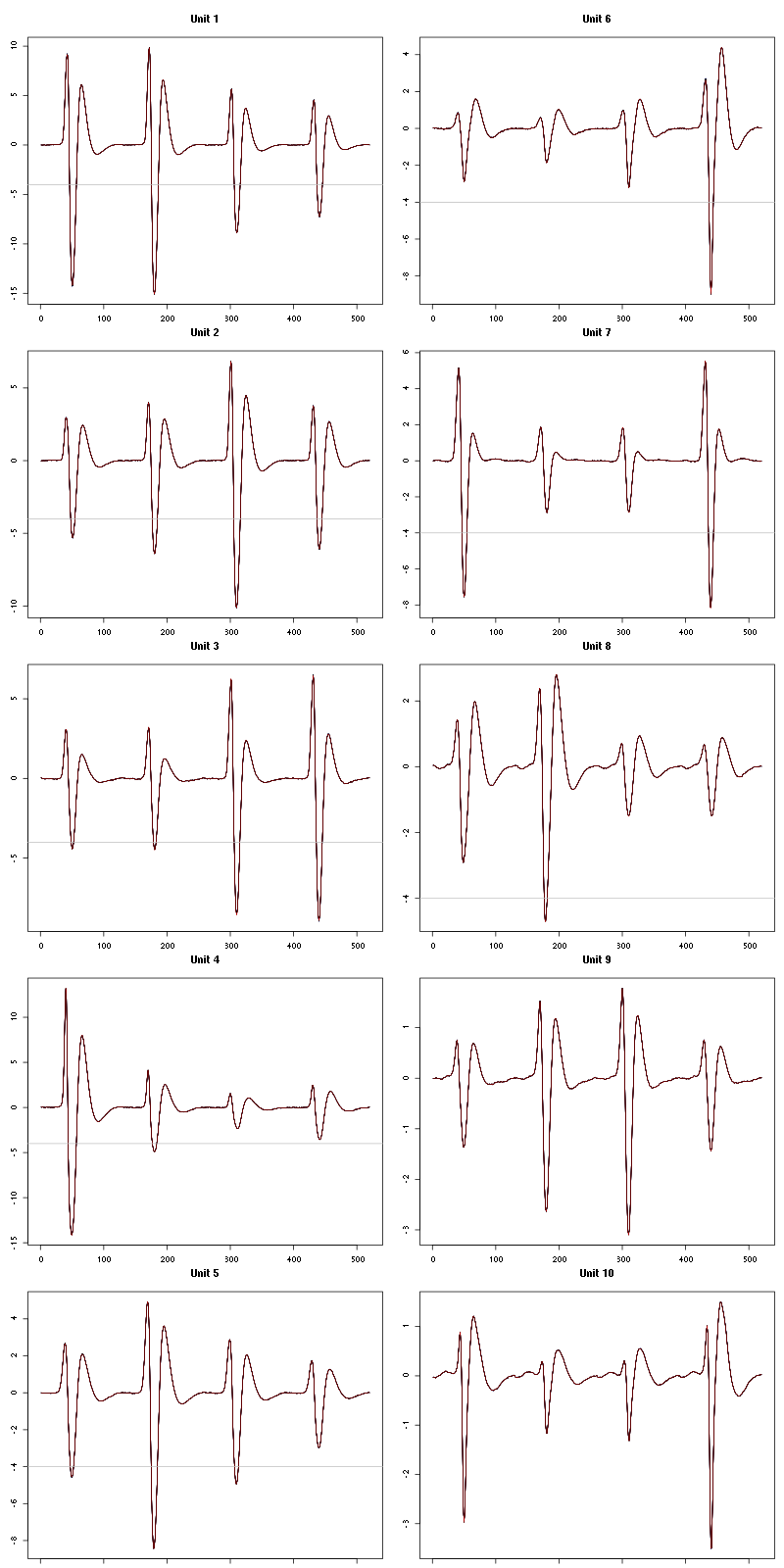
Figure 58: Evolution of the templates of each unit during the 10 trials of C3H_4 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
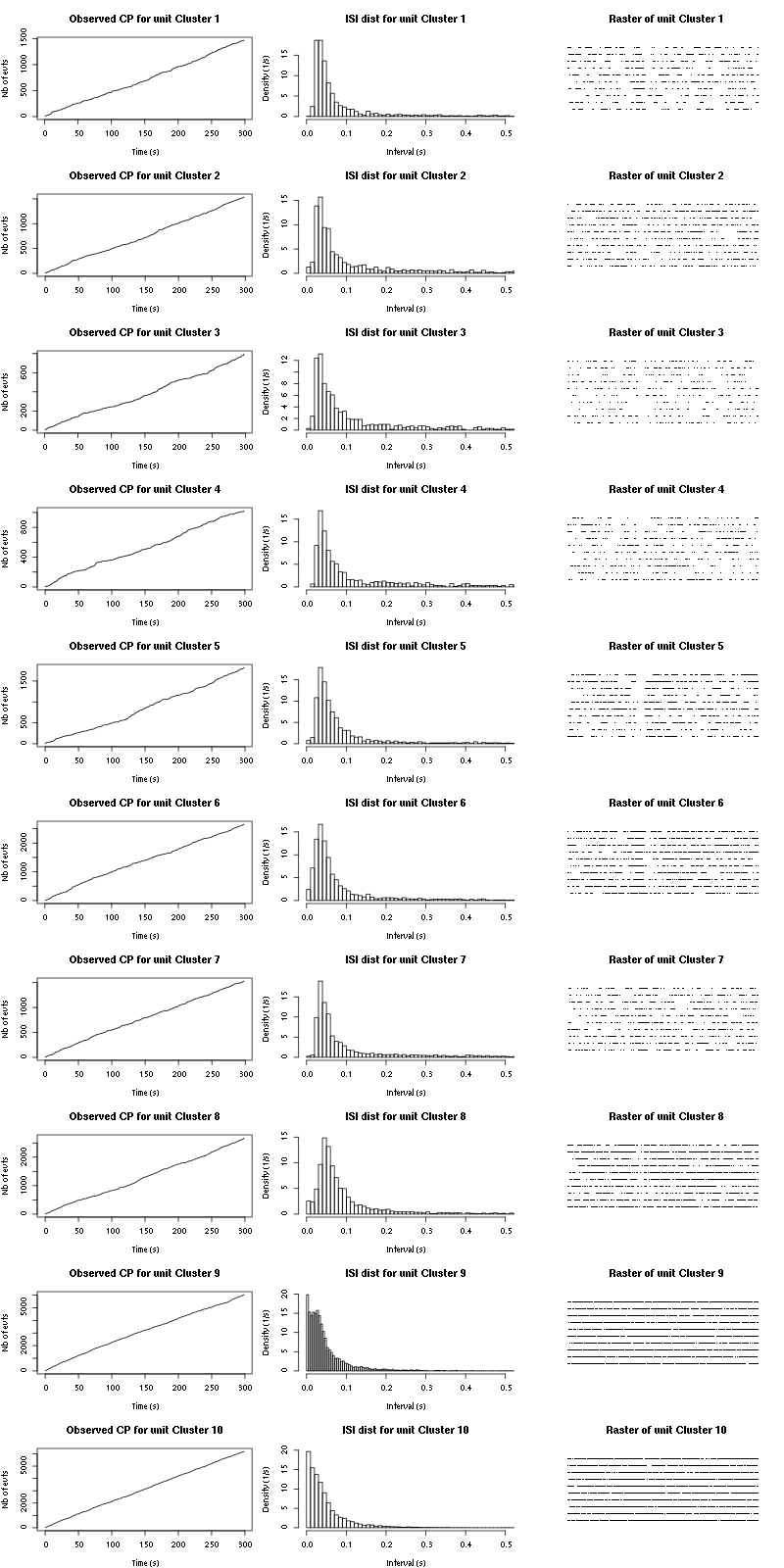
Figure 59: Observed counting processes, empirical inter spike interval distributions and raster plots for C3H_4.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_4_tetB$spike_trains[[i]],
a_C3H_4_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
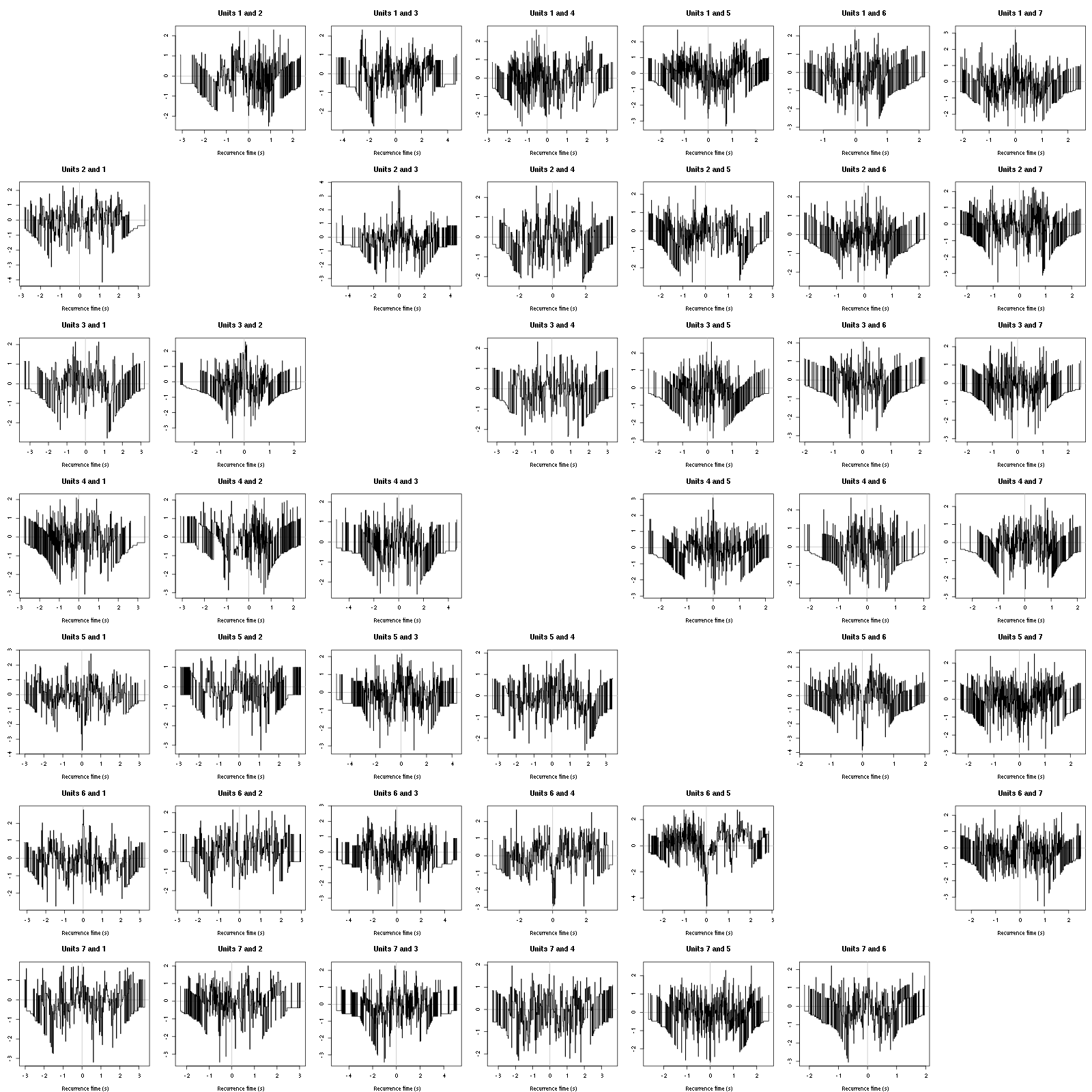
Figure 60: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
15.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_4_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_4_tetB$spike_trains))
if (!is.null(a_C3H_4_tetB$spike_trains[[c_idx]]))
cat(a_C3H_4_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_C3H_4_tetB_u",c_idx,".txt"),sep="\n")
16 30 trials of C3H_5
The LabBook mentions noise in trials 21, 22 and 23 so we do not consider them.
16.1 Do the job
stim_name = "C3H_5"
a_C3H_5_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=c(1:20,24:30),
centers=a_C3H_4_tetB$centers,
counts=a_C3H_4_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
16.2 Diagnostic plots
The counts evolution is:
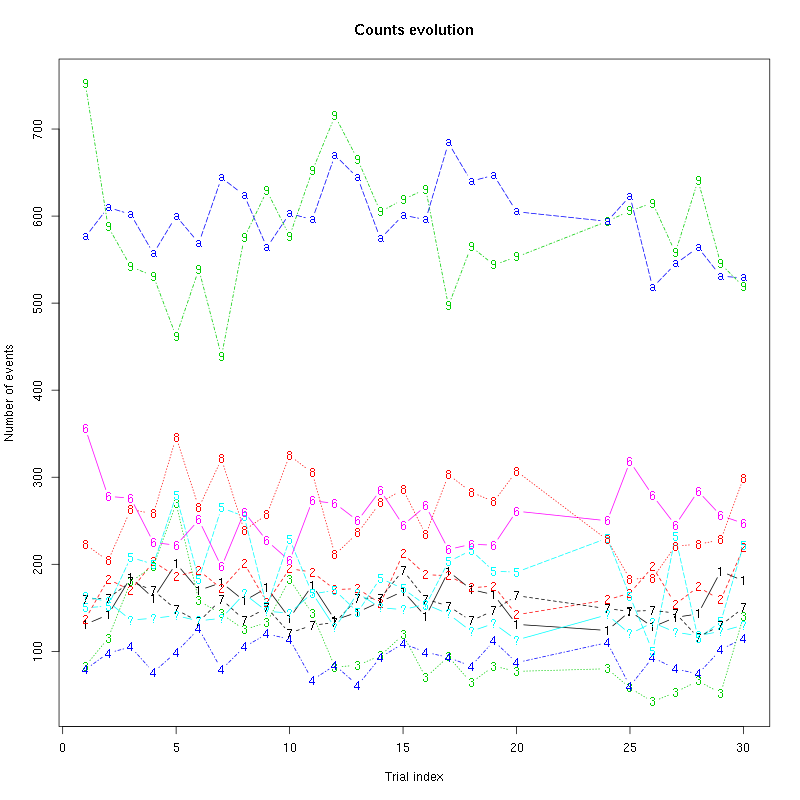
Figure 61: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 27 trials of C3H_5 for tetrodeB.
The waveform evolution is:
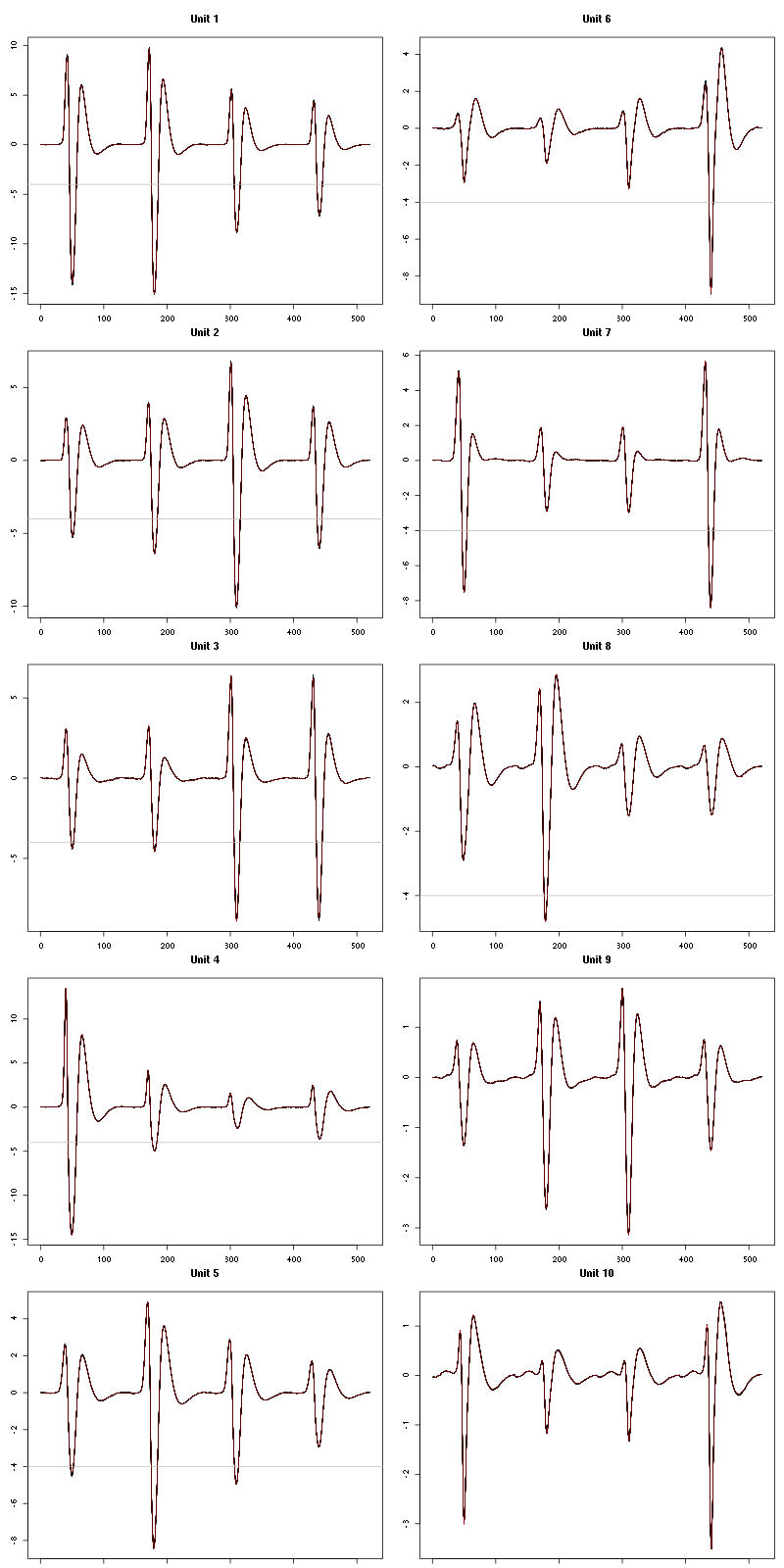
Figure 62: Evolution of the templates of each unit during the 27 trials of C3H_5 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
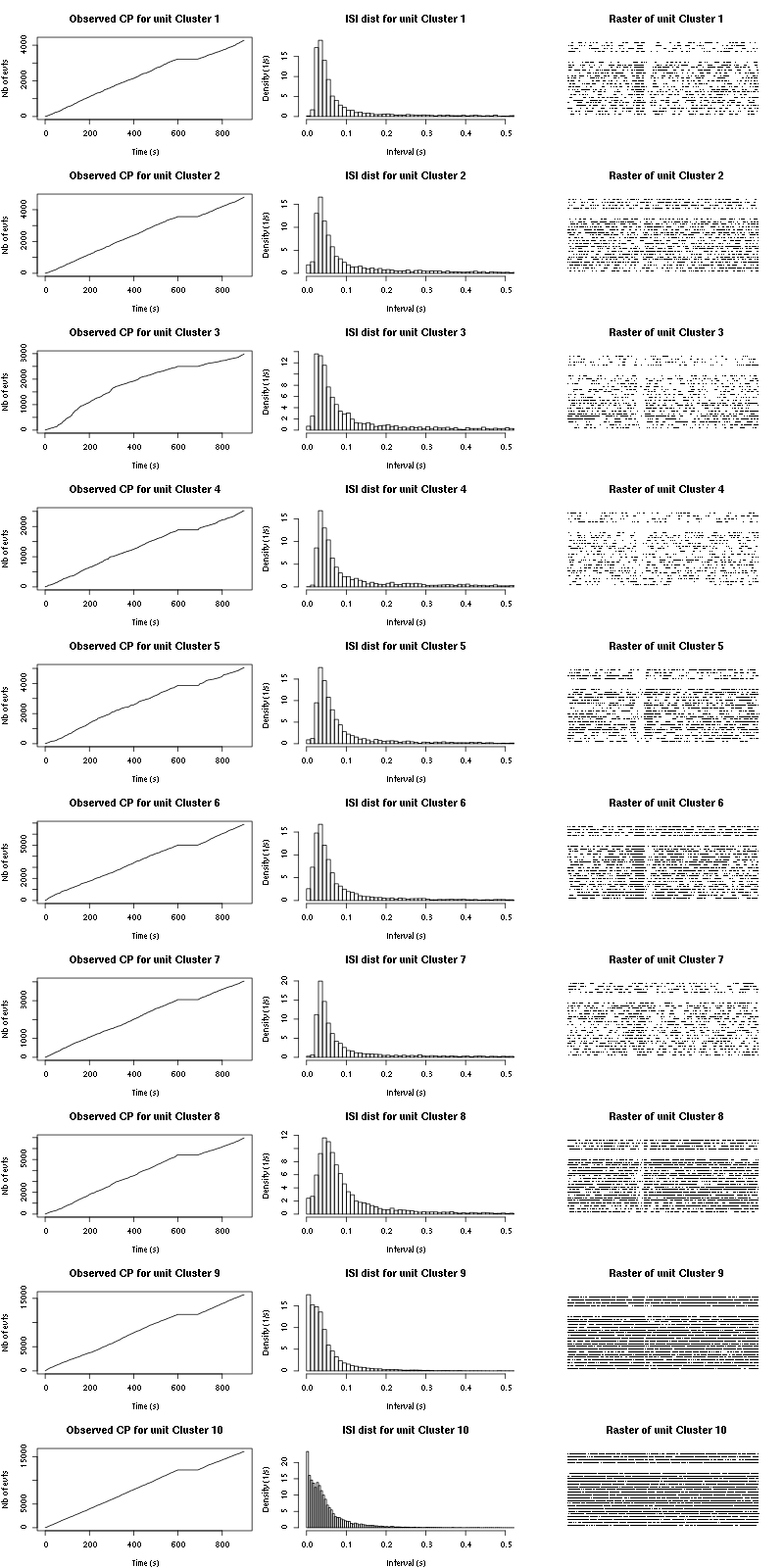
Figure 63: Observed counting processes, empirical inter spike interval distributions and raster plots for C3H_5.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_5_tetB$spike_trains[[i]],
a_C3H_5_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
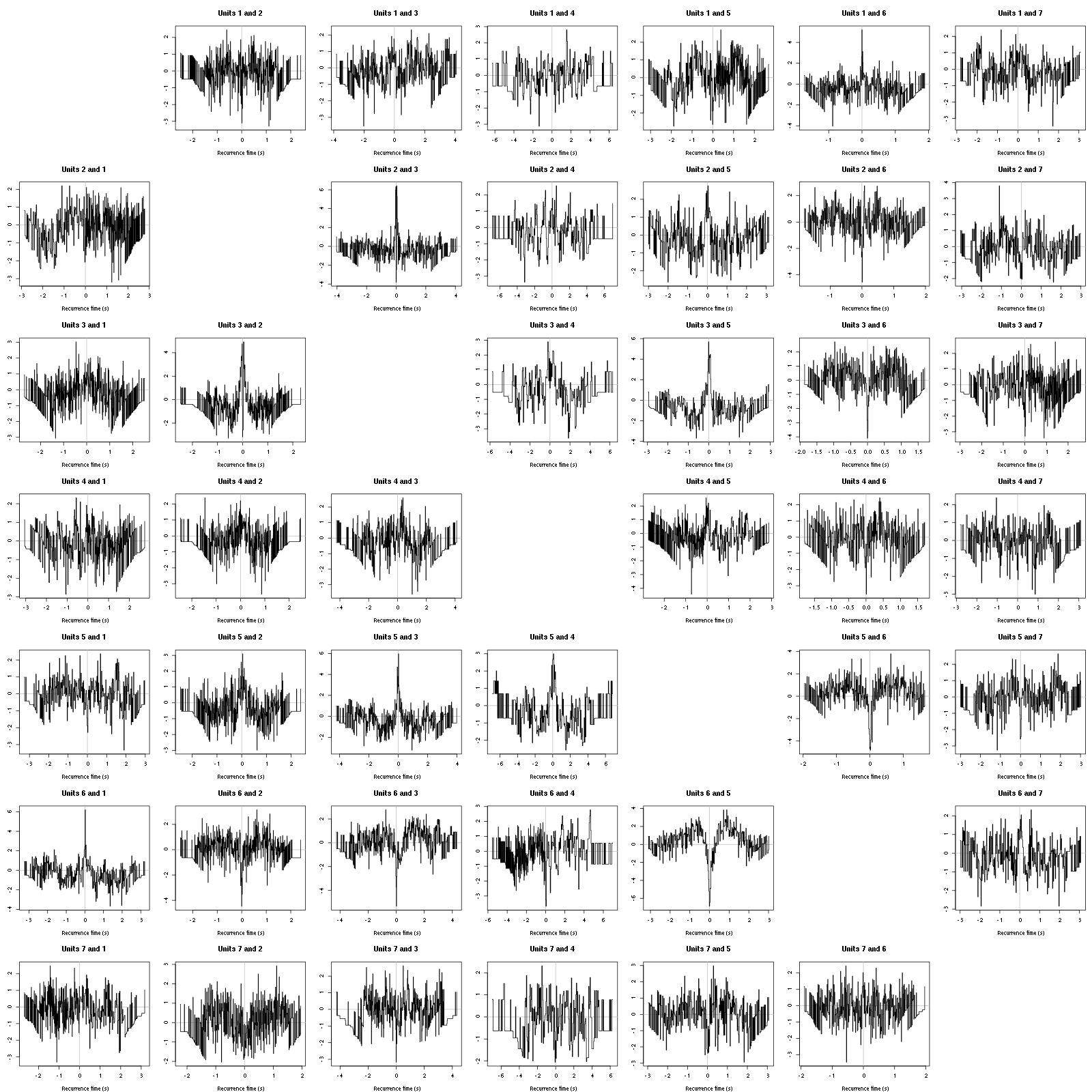
Figure 64: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
16.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_5_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_5_tetB$spike_trains))
if (!is.null(a_C3H_5_tetB$spike_trains[[c_idx]]))
cat(a_C3H_5_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_C3H_5_tetB_u",c_idx,".txt"),sep="\n")
17 30 trials of C3H_6
17.1 Do the job
stim_name = "C3H_6"
a_C3H_6_tetB=sort_many_trials(inter_trial_time=30*15000,
get_data_fct=get_tetrode_data,
stim_name=stim_name,
trial_nbs=1:30,
centers=a_C3H_5_tetB$centers,
counts=a_C3H_5_tetB$counts,
all_at_once_call_list=list(thres=threshold_factor*c(1,1,1,1),
filter_length_1=filter_length, filter_length=filter_length,
minimalDist_1=15, minimalDist=15,
before=c_before, after=c_after,
detection_cycle=c(0,0), verbose=1),
layout_matrix=matrix(c(1,1:11),nr=6),new_weight_in_update=0.01
)
17.2 Diagnostic plots
The counts evolution is:
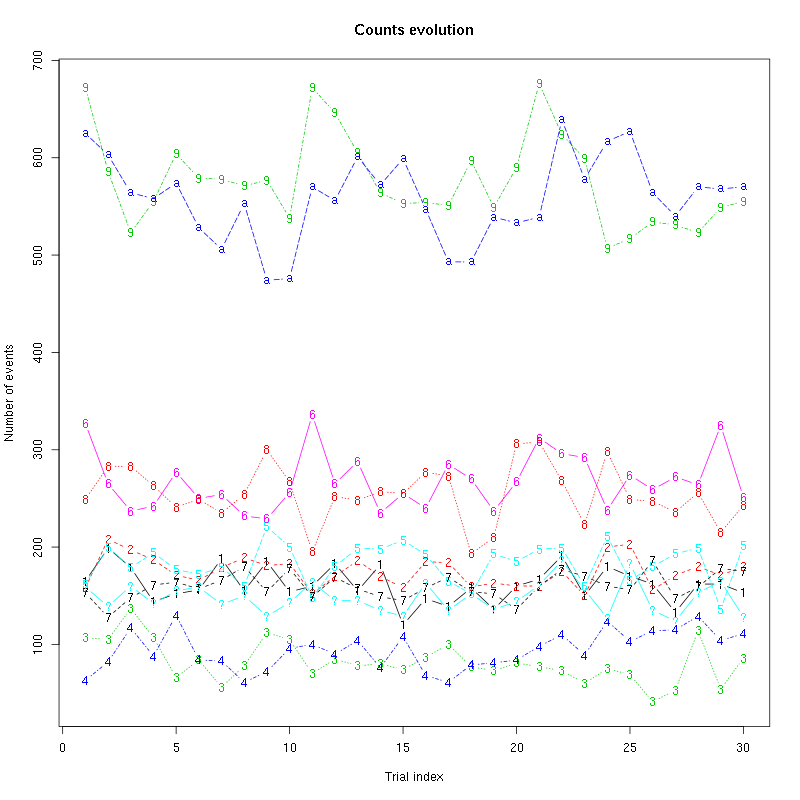
Figure 65: Evolution of the number of events attributed to each unit (1 to 9 and "a") or unclassified ("?") during the 30 trials of C3H_6 for tetrodeB.
The waveform evolution is:
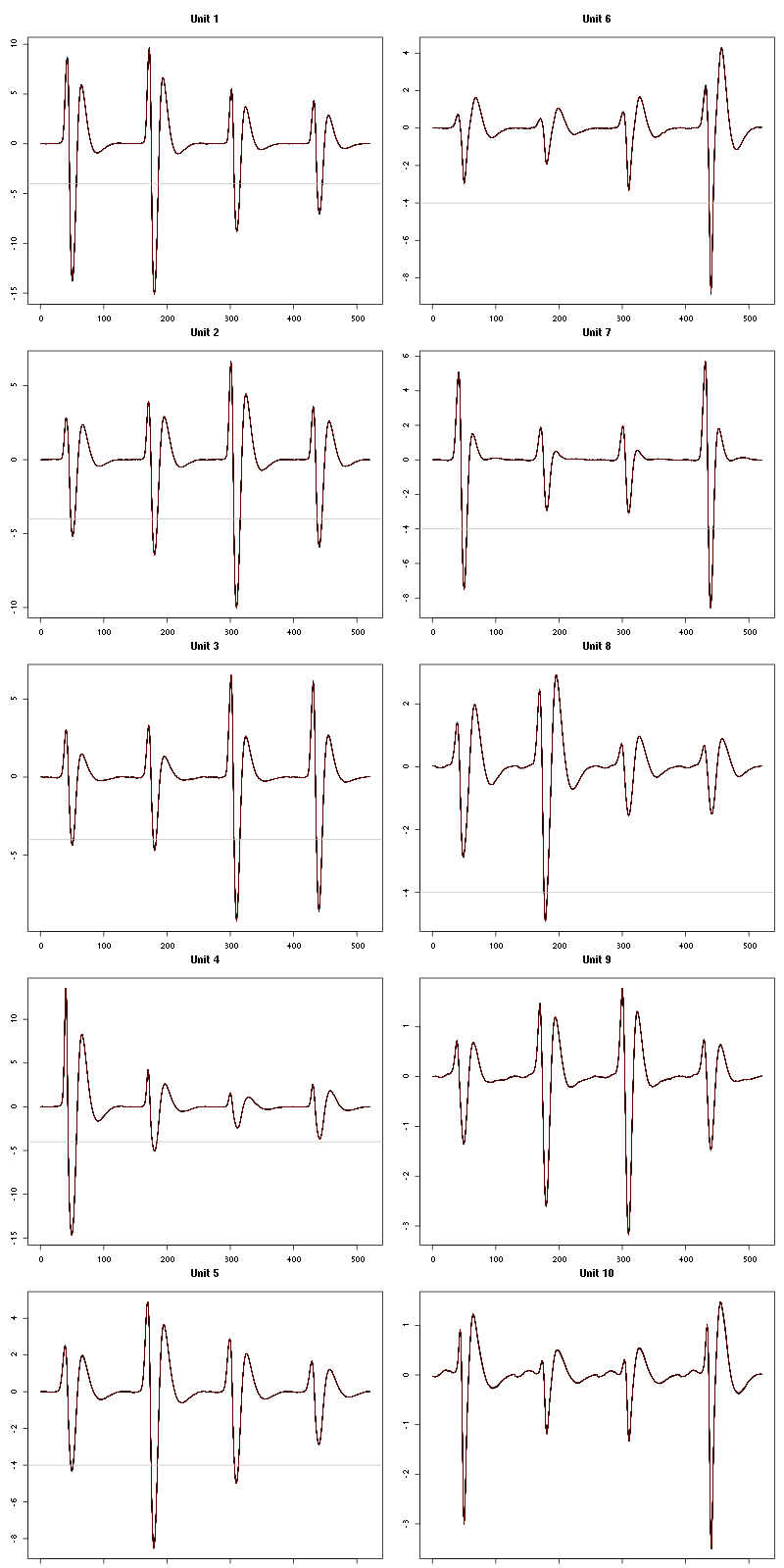
Figure 66: Evolution of the templates of each unit during the 30 trials of C3H_6 for tetrodeB.
The observed counting processes and inter spike intervals densities are:
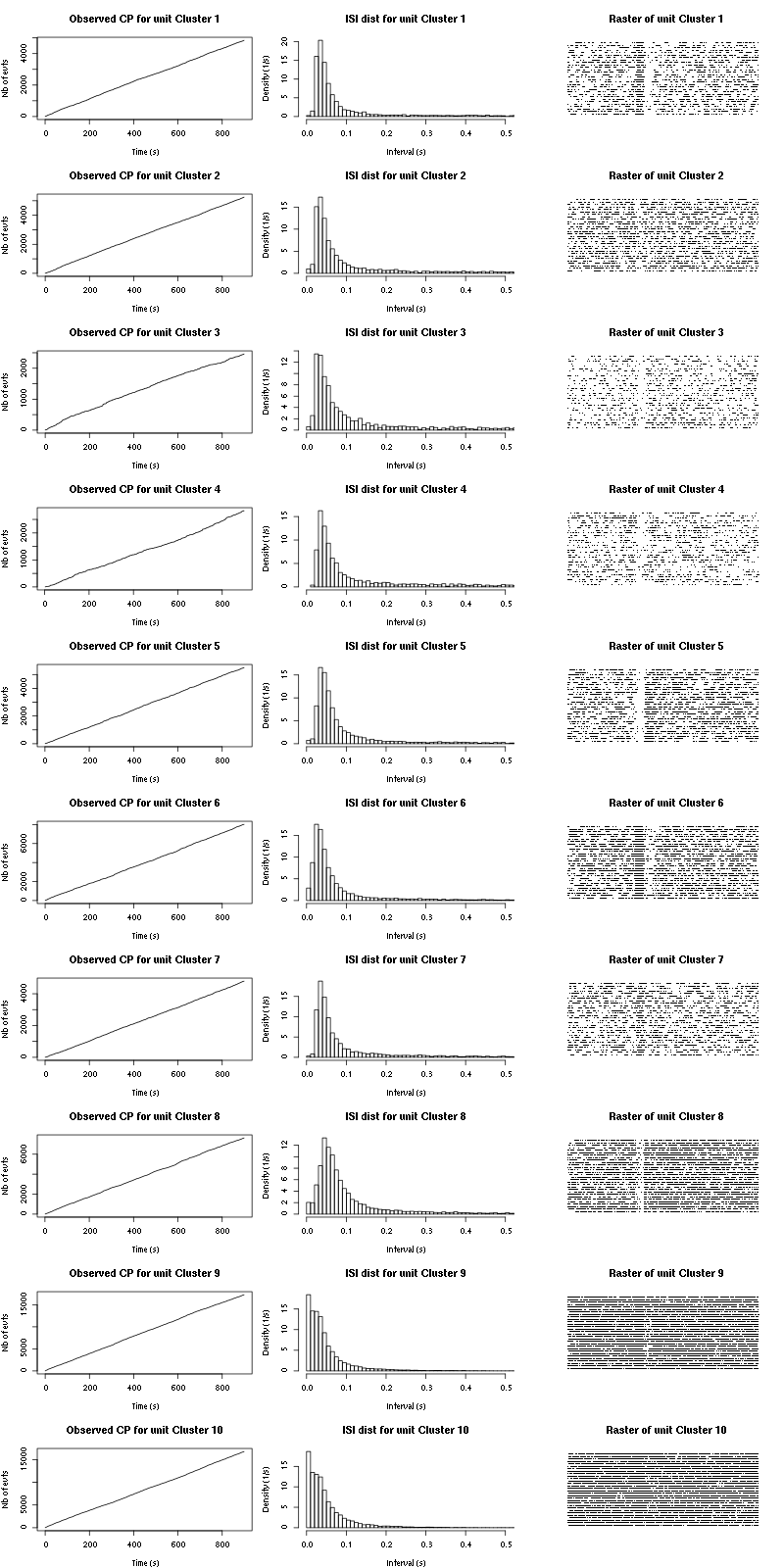
Figure 67: Observed counting processes, empirical inter spike interval distributions and raster plots for C3H_6.
The BRT and RFT tests give for the first 7 units:
layout_matrix = matrix(0,nr=nbc-3,nc=nbc-3)
counter = 1
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j) {
layout_matrix[i,j] = counter
counter = counter +1
}
layout(layout_matrix)
par(mar=c(4,3,4,1))
for (i in 1:(nbc-3))
for (j in 1:(nbc-3))
if (i != j)
test_rt(a_C3H_6_tetB$spike_trains[[i]],
a_C3H_6_tetB$spike_trains[[j]],
nbins=200,single_trial_duration=30,
ylab="",main=paste("Units",i,"and",j))
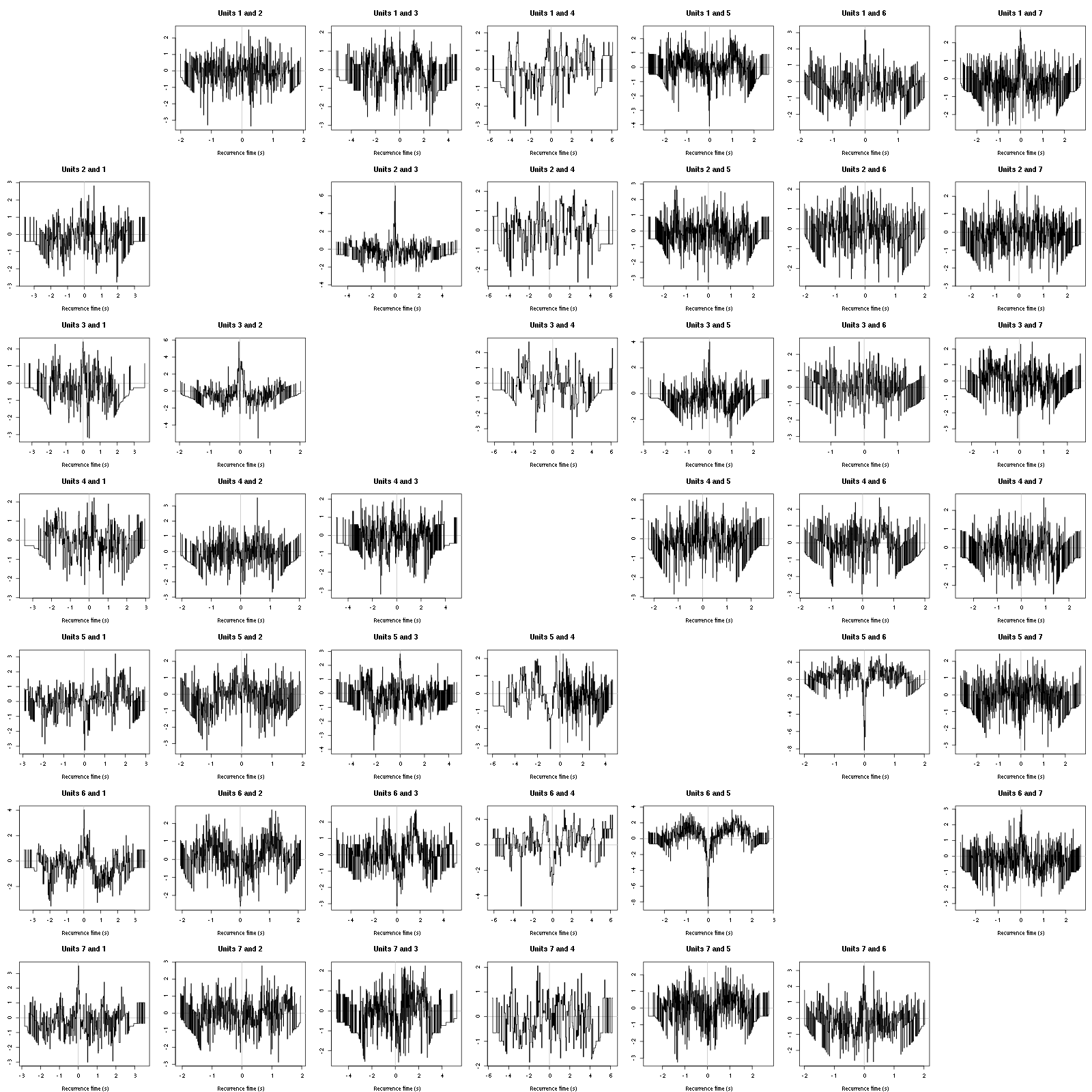
Figure 68: Graphical tests for the first 7 units of the Backward and Forward Reccurrence Times distribution against the null hypothesis (no interaction) during Spontaenous_1. If the null is correct, the curves should be IID draws from a standard normal distribution.
17.3 Save results
Before analyzing the next set of trials we save the output of sort_many_trials to disk with:
save(a_C3H_6_tetB,
file=paste0("tetB_analysis/tetB_",stim_name,"_summary_obj.rda"))
We write to disk the spike trains in text mode:
for (c_idx in 1:length(a_C3H_6_tetB$spike_trains))
if (!is.null(a_C3H_6_tetB$spike_trains[[c_idx]]))
cat(a_C3H_6_tetB$spike_trains[[c_idx]],file=paste0("locust20010214_spike_trains/locust20010214_C3H_6_tetB_u",c_idx,".txt"),sep="\n")